Fig. 1.
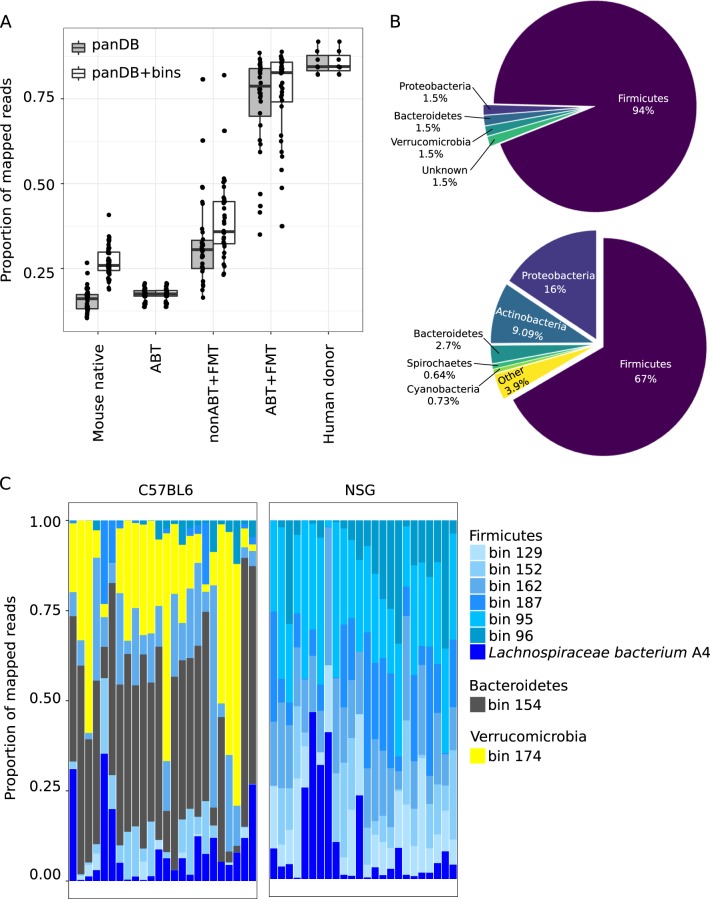
Reconstruction of the mouse gut microbiome using a combination of reference-based and de novo profiling. a the proportion of reads mapped to the reference pan-genomes in the database panDB and high-quality genome bins reconstructed de novo from native mouse gut metagenomic reads. b Phylum-level taxonomic distribution of the high-quality genome bins (upper) and assembled contigs that were not assigned into the genome bins (lower). c Mouse gut metagenomic reads mapped to the most abundant species. Here, “species” refers to both microbial species in panDB and the high-quality genome bins. Each column corresponds to one mWGS sample, the proportion of reads mapped to the most abundant taxa shown in the graph were rescaled so that they added up to 1