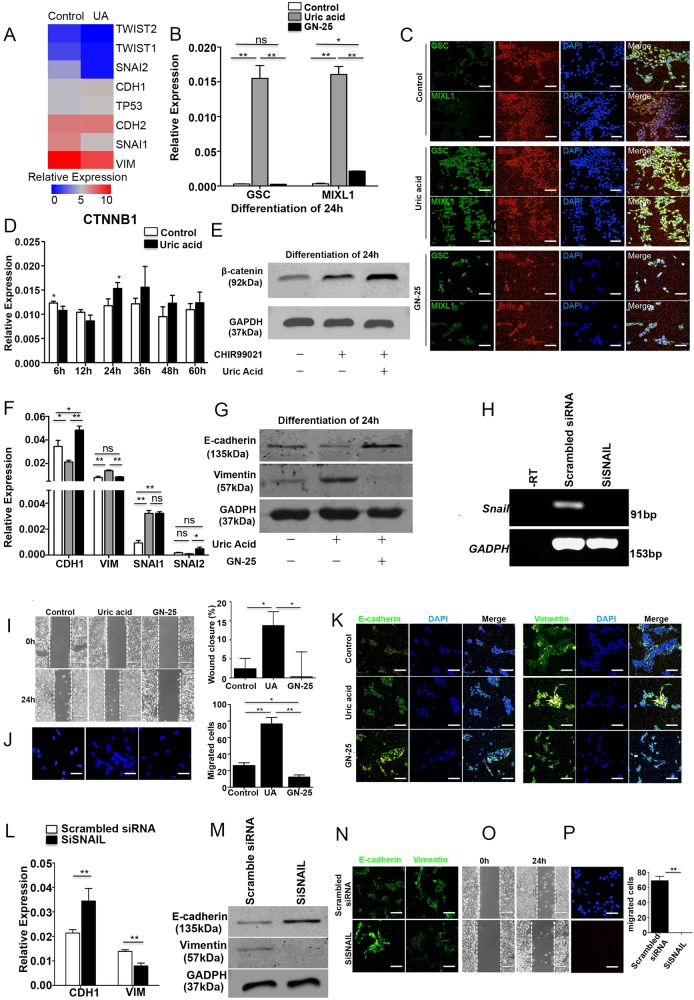
Fig. 6.
UA promoted cardiac differentiation through increasing EMT. a Heatmap shows hierarchical clustering of EMT genes. b The expression of mesoderm genes, GSC and MIXL1, were examined with quantitative PCR with or without UA and GN-25. The results were expressed as relative expression to GADPH and plotted as percentages of the maximum. c Immunostaining of GSC or MIXL1 and BrdU in 24 h. Scale bars = 200 μm. d The expression of CTNNB1 was examined with quantitative PCR with or without UA for every 12 h. The results were expressed as relative expression to GADPH and plotted as percentages of the maximum. e Immunoblot analysis of beta-catenin in 24 h. The results were expressed as relative expression to GADPH. f The expression of EMT genes, VIM, CDH1 SNAI1, and SNAIL2 were examined with quantitative PCR with or without UA and GN-25. The results were expressed as relative expression to GADPH and plotted as percentages of the maximum. g Immunoblot analysis of e-cadherin and vimentin in 24 h. The results were expressed as relative expression to GADPH. h Gel electrophoresis with SNAIL1 SiRNA. The results were expressed as relative expression to GADPH. (i) NKX2–5-GFP hESCs with or without UA treatment was analyzed in an in vitro wound model. Scale bars = 200 μm. The percent wound closure (%) was counted under a light microscope. j Transwell assay in vitro. Migration cell was counted by DAPI in fluorescence microscope. Scale bars = 200 μm. k Immunostaining of e-cadherin and vimentin with or without UA and GN-25 for 24 h. Scale bars = 200 μm. l The expression of EMT genes, VIM and CDH1, were examined with quantitative PCR with SiRNA. The results were expressed as relative expression to GADPH and plotted as percentages of the maximum. m Immunoblot analysis of e-cadherin and vimentin. The results were expressed as relative expression to GADPH. n Immunostaining of e-cadherin and vimentin with SNAIL1 SiRNA. Scale bars = 200 μm. o NKX2–5-GFP hESCs was transfected with SiRNA and analyzed in an in vitro wound model. Scale bars = 200 μm. The percent wound closure (%) was counted under a light microscope. p Transwell assay in vitro. Migration cell was counted by DAPI in fluorescence microscope. Scale bars = 200μm. n = 3 each. Data are expressed as means ± SD. *P < 0.05, **P < 0.01 vs. control