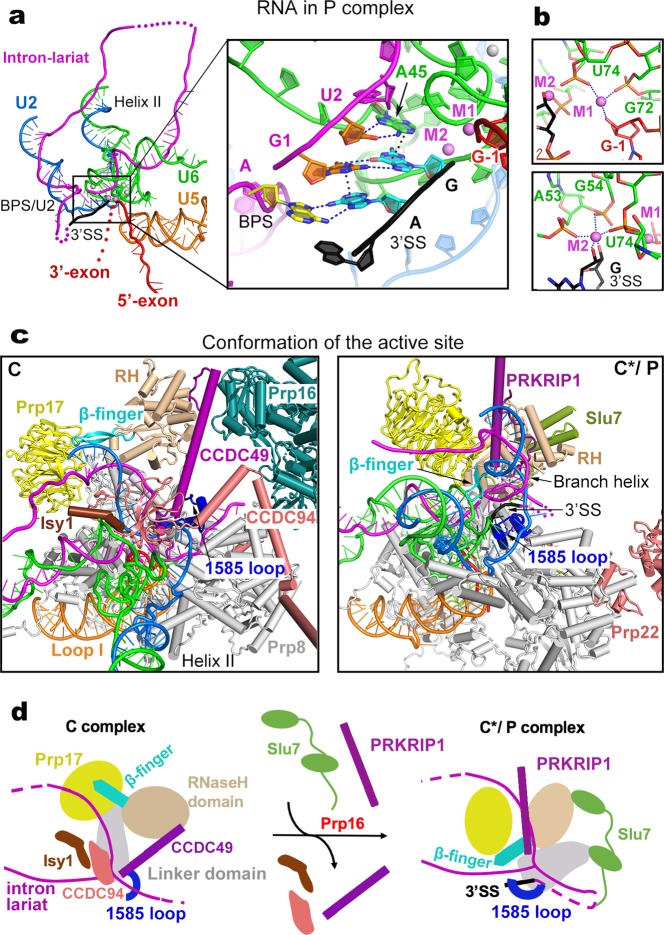
Fig. 2.
Structural features of the human P complex. a Structure of the RNA elements in the P complex. An overall cartoon representation of the RNA map is shown on the left panel, and a close-up view on the splicing active site center is displayed in the right panel. Disordered RNA sequences in the intron lariat and the 3′-exon are represented by magenta and red dotted lines, respectively. The catalytic and structural metals are shown in magenta and gray spheres, respectively. The 3′-splice site (3′SS) is highlighted in black. Putative hydrogen bonds and van der Waals interactions that mediate recognition of the 3′SS are represented by blue and black dotted lines, respectively. b Specific coordination of the catalytic metal ions in the human P complex. c Structural rearrangements of the spliceosomal components at the active site during the C-to-P transition. The human C complex (left panel) and P complex (right panel) are shown in the same orientation as determined by the core of Prp8 and U5 snRNA. Shown here are the proteins and RNA elements around the catalytic center. d A cartoon diagram of the 3′SS recognition. Remodeling of the C complex by Prp16 results in dissociation of the NTC component Isy1 and the step I factors CCDC49 and CCDC94/YJU2. The Linker domain of Prp8 is rotated away from the U2/BPS duplex. Consequently, the 1585-loop which binds the 3′-tail of the intron loads the 3′SS into the splicing active site center. The RNaseH-like domain is translocated towards the active site, pushing the β-finger towards the lariat junction. The WD40 domain of Prp17 is also translocated toward the branch helix; together with the splicing factor PRKRIP1 stabilizes the new conformation of RNaseH-like domain and branch helix. The intron lariat junction is sandwiched by the β-finger and the 1585-loop. Slu7 adopts an extended conformation and binds the RNaseH-like and Linker domains of Prp8, stabilizing the local conformation