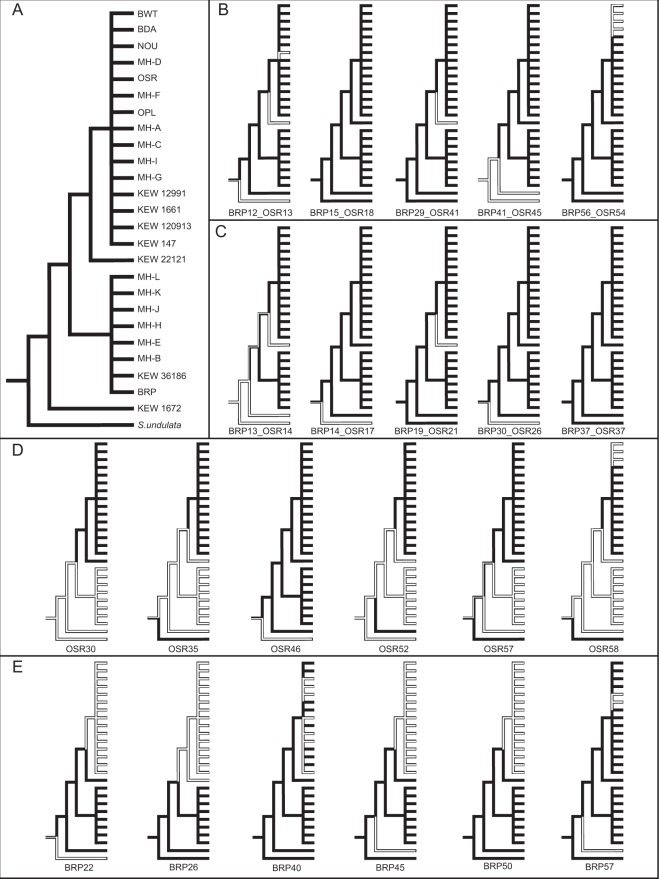
Fig. 4.
Parsimony-based reconstruction of ancestral States for the presence (black) or absence (white) of each of the 22 sampled chromosomes across 25 S. noctiflora samples and the close relative S. undulata. a The constraint topology used for the analysis is based on concatenated sequence data from the mitochondrial genome (see Fig. 1a), and placement of the multiple OSR-like and BRP-like samples is their respective groups in based on the phylogenetic analysis in Figure S1. The remaining panels show the presence/absence states for b the five sampled chromosomes with high sequence divergence, c the five sampled chromosomes that are conserved between BRP and OSR and have no genes, d the six sampled “OSR-specific” chromosomes, and e the six sampled “BRP-specific” chromosomes