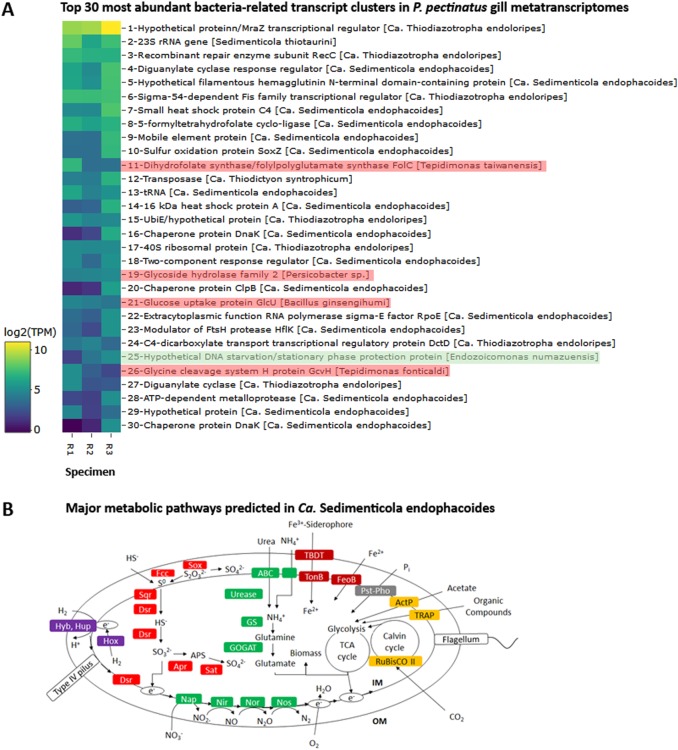
Fig. 3.
Log2-transformed TMM-normalized TPM of gene products of a the 30 most abundantly expressed protein-coding transcript clusters mapped to species (square brackets) from the domain Bacteria and b morphological features and major metabolic pathways predicted in Ca. Sedimenticola endophacoides. In b, the transcript cluster mapped to a non-thioautotrophic gammaproteobacterial species (Endozoicomonas numazuensis) is highlighted in green while transcript clusters mapped to non-gammaproteobacterial taxa are highlighted in pink. UbiE ubiquinone/menaquinone biosynthesis C-methyltransferase, FtsH ATP-dependent zinc metalloprotease, Hyb membrane bound [Ni-Fe] hydrogenase 2, Hup uptake hydrogenase, Hox soluble NAD-dependent hydrogenase, S0 elemental sulfur, Fcc flavocytochrome c-sulfide dehydrogenase, Sqr sulfide:quinone oxidoreductase, Sox sulfur oxidation enzyme complex, Dsr reverse dissimilatory sulfite reductase enzyme system, Apr adenylylsulfate reductase, APS adenosine-5’-phosphosulfate, Sat sulfate adenylyltransferase, ABC ATP-binding cassette transporters, GS glutamine synthetase, GOGAT glutamine oxoglutarate aminotransferase (glutamate synthase), Nap periplasmic dissimilatory nitrate reductase, Nir cytochrome nitrite reductase cd1, Nor nitric oxide reductase, Nos nitrous oxide reductase, TBDT TonB-dependent transporter, TonB TonB-ExbB-ExbD complex, FeoB ferrous iron transport protein, Pst phosphate specific transport, Pho phosphate regulon, PolyP polyphosphate granule, ActP acetate permease, TRAP tripartite ATP-independent periplasmic transport, RuBisCO ribulose-1,5-bisphosphate carboxylase/oxygenase, TCA cycle tricarboxylic acid cycle, IM inner membrane, OM outer membrane