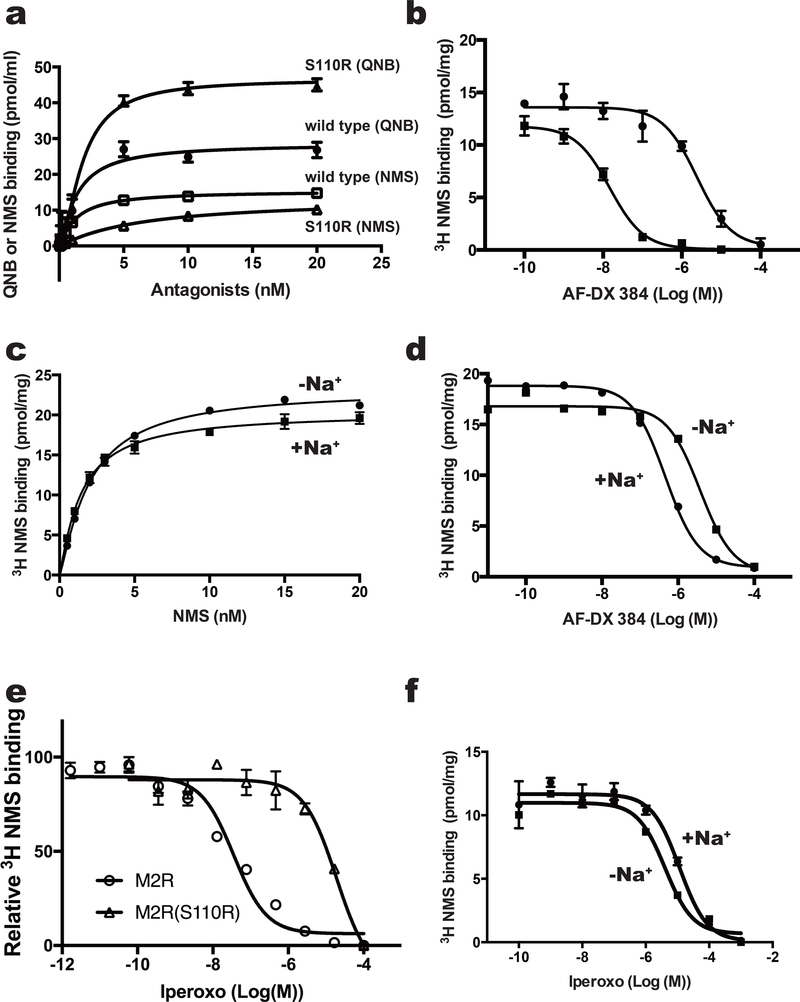
Figure 2.
Binding assay of M2 variants with agonist and antagonists. (a) Effects of mutation at position 3.39 on equilibrium binding of the indicated radioligands. Saturation curves for binding of the inverse agonist QNB to M2-BRIL (filled circles) or the S110R mutant (filled triangles), and of the inverse agonist NMS to M2-BRIL (open squares) or the S110R mutant (open triangles). (b) Inhibition of [3H]NMS binding with M2-BRIL (filled circles) and the S110R mutant (filled square) by AF-DX 384. (c and d) Analysis of sodium ion–dependent binding of (c) NMS and (d) AF-DX 384 to M2-BRIL. (e) Inhibition of [3H]NMS binding with the M2 receptor_delta_ICL3 (open circle) and S110R mutant (open square) by the agonist iperoxo. (f) Inhibition of [3H]NMS binding with the M2-BRIL by the agonist iperoxo with (closed circle) or without (closed square) sodium ions. In a and c, error bars, s. e. m. (n = 3 separate binding assays on the same cell-membrane stock). In b, d, e and f, error bars, s. e. m. (n = 3 separate competition assays on the same cell-membrane stock).