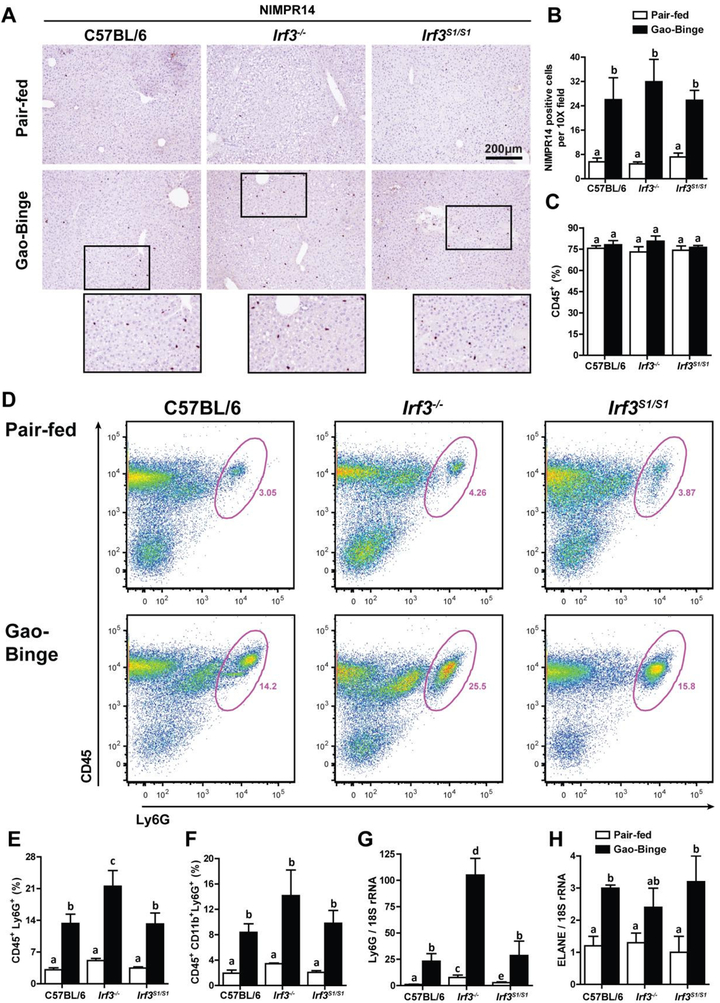
Figure 4: Neutrophil accumulation in liver in response to Gao-binge ethanol exposure was independent of Irf3 genotype.
C57BL/6, Irf3−/− and Irf3S1/S1 mice were exposed to the Gao-binge model as in Figure 1. A/B) NIMPR14 expression was visualized by immunohistochemistry after 6 h. A) Formalin-fixed paraffin-embedded sections of liver were de-paraffinized and expression of NIMPR14 assessed by immunohistochemistry. B) Images were acquired at 10X and the number of positive cells enumerated. C/D/E/F) Total non-parenchymal cells were isolated from the liver and analyzed by flow cytometry. The percentage of C) CD45+, D/E) CD45+/Ly6G+ and F) CD45+/ CD11b+/Ly6G+ was determined by flow cytometry. D) Representative flow diagrams for CD45+/Ly6G+ are illustrated. G/H) Expression of G) Ly6G and H) ELANE mRNA in whole liver was assessed by qRT-PCR and normalized to 18S rRNA. Values represent means ± SEM, n=4–8 per group. Values with different superscripts are significantly different from each other, p<0.05, assessed by ANOVA..