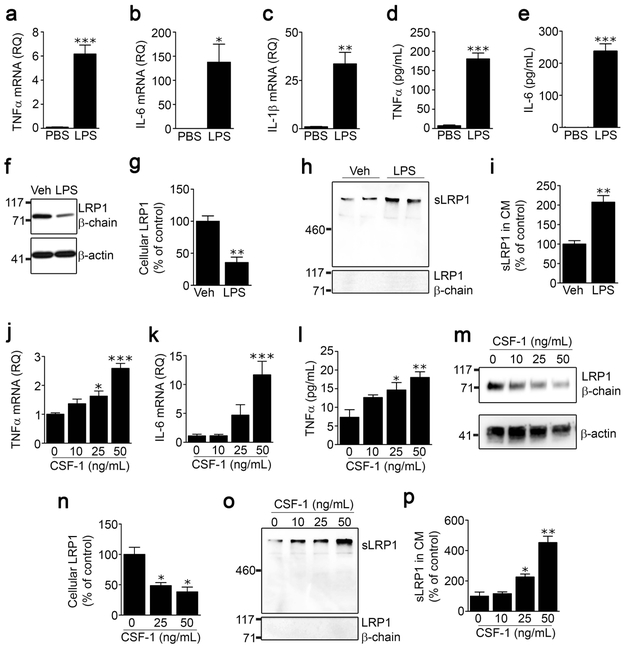
FIGURE 2.
LPS and CSF-1 induce LRP1 shedding from spinal cord microglia. Microglia were isolated from spinal cords of adult C57BL/6J wild-type mice and cultured in low-serum medium. The cells were treated with LPS (100 ng/mL) or vehicle for 24 h. Expression of (a) TNFα, (b) IL-6 and (c) IL-1β was determined by RT-qPCR. (d, e) TNFα protein and IL-6 protein in CM were measured by ELISA (mean ± s.e.m. of 4 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t-test). (f) Cell extracts were subjected to immunoblot analysis to detect LRP1 β-chain (upper panel) and β-actin as a loading control (lower panel). (g) Densitometry was performed to compare cellular LRP1 in microglia before and after LPS treatment (mean ± s.e.m; n = 3 independent immunoblotting experiments; **p < 0.01, two-tailed unpaired t-test) (h) Conditioned medium was collected and subjected to RAP ligand-blotting to detect sLRP1 (upper panel). The same samples were subjected to immunoblot analysis using an antibody that detects an epitope in the LRP1 β-chain, absent in sLRP1 (lower panel). These immunoblots were performed concurrently with those shown in panel “f’ of this figure, providing a positive control. (i) Densitometry was performed to quantify sLRP1 in CM (mean ± s.e.m; n = 4 independent experiments; **p < 0.01, two-tailed unpaired t-test). (j, k) Microglia were treated with increasing concentrations of mouse CSF-1. RNA was isolated 24 h later and RT-qPCR was performed to determine expression of TNFα and IL-6. (l) CM was recovered. TNFα protein was determined by ELISA (mean ± s.e.m.; n = 3 independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA followed by Dunnett’s post-hoc test). (m, n) Microglia were treated with the indicated concentrations of CSF-1. Cellular LRP1 was determined by immunoblot analysis using anti LRP1 β-chain antibody and quantified by densitometry (mean ± s.e.m.; n = 3 independent experiments; *p < 0.05, one-way ANOVA followed by Dunnett’s post-hoc test). (o) sLRP1 released into CM was detected by RAP ligand-blotting. The same samples were subjected to immunoblot analysis using LRP1 β-chain antibody. (p) Densitometry was performed to quantify sLRP1 in CM (mean ± s.e.m.; n = 3 independent experiments; *p < 0.05, **p < 0.01, one-way ANOVA followed by Dunnett’s post-hoc test).