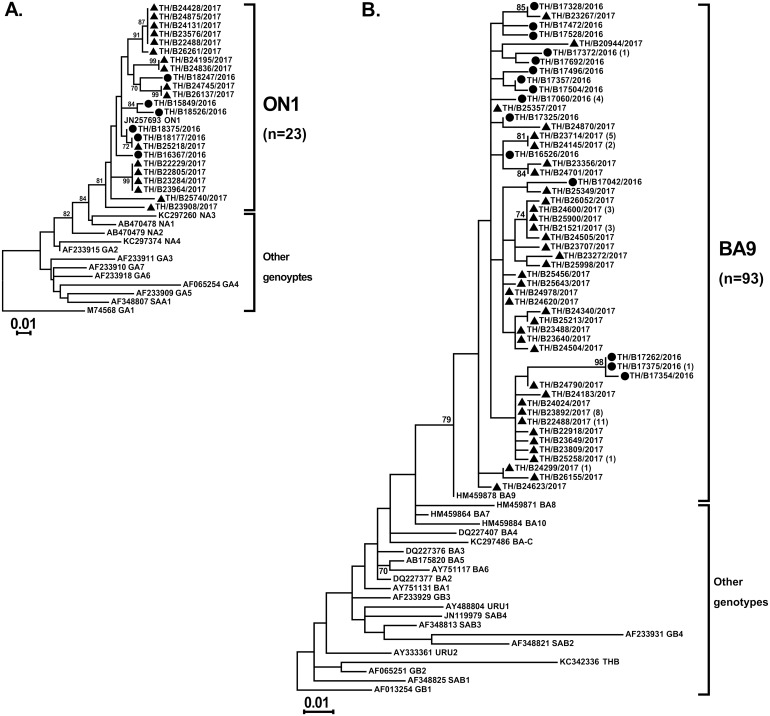
Figure 2. Phylogenetic analysis of RSV subgroup A and B based on the nucleotide sequence encompassing the HVR2 region within the G gene.
Trees were constructed using the maximum likelihood method based on the Tamura–Nei model and implemented in MEGA6. Bootstrap values of 1,000 pseudo-replications >70% are indicated at the branch nodes. Reference sequences for each genotype (GA1–GA7, SAA1, NA1–NA4, and ON1 for RSV-A and GB1–GB4, SAB1–SAB4, URU1, URU2, THB, and BA1–BA10 for RSV-B) were obtained from GenBank. The scale bar represents the number of nucleotide substitutions per site between close relatives. Circles denote samples from Thailand 2016, while squares indicate strains from Thailand 2017 The number of strains are shown in parentheses.