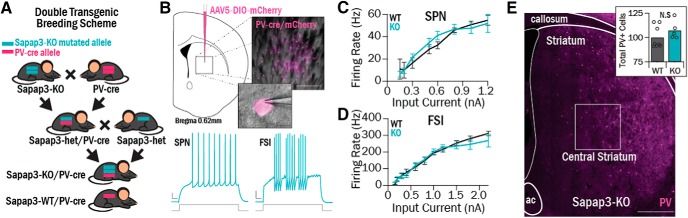
Figure 1.
Central striatum cell types were interrogated using ex vivo electrophysiology in double-transgenic Sapap3-KO//PV-cre mice. A, Double-transgenic mice were generated to investigate functional properties of FSIs. Sapap3-KO mice were bred with PV-cre mice to generate mice with one Sapap3-KO allele and one PV-cre allele (Sapap3-het//PV-Cre). Sapap3-het//PV-Cre mice were then used as breeders to generate Sapap3-KO//PV-cre and Sapap3-WT//PV-cre progeny. B, Top, AAV5-DIO-mCherry was injected in central striatum; resulting infection of cre-positive PV cells led to fluorescent labeling for targeted slice electrophysiology recordings. Scale bars: Inset top, 200 μm; Inset bottom, 10 μm. Bottom, Examples of evoked spiking traces in KO SPNs and FSIs (right). Calibration: 50 ms, 10 mV. C, No differences were observed between SPN I-O curves in Sapap3-KOs (4 animals, 25 cells) and WTs (4 animals, 14 cells). D, No differences were observed between FSI I-O curves in Sapap3-KOs (3 animals, 23 cells) and WTs (5 animals, 18 cells). E, Tissue sections from Sapap3-KO (N = 5) and WT (N = 8) mice were immunohistochemically stained for PV to perform FSI cell counts in central striatum (box). Scale bar, 500 μm. Inset, There was no difference observed in the number of FSIs in WTs versus KOs (rank sum = 44, p = 0.21, WRST).