Figure 1.
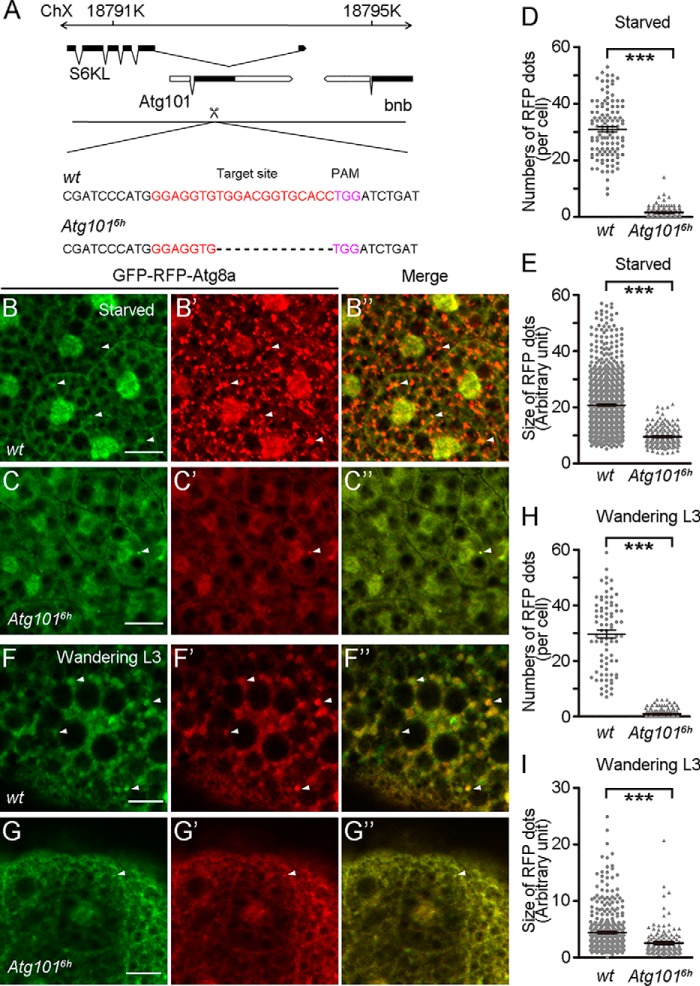
Loss of Atg101 causes defects in starvation-induced and developmental autophagy. A, schematic diagram for the Atg101 gene locus. Coding exons are in black, and noncoding exons are in white. The guide RNA target sequence is indicated in red. The PAM sequence is indicated in purple. The dotted line indicates the deleted sequence. Atg1016h has a 13-bp deletion around the gRNA target site. ChX, chromosome X. B–B″, accumulation of GFP-Atg8a and RFP-Atg8a punctate structures in fat body cells of middle stage WT third instar larvae in response to starvation. C–C″, absence of GFP-Atg8a and RFP-Atg8a punctate structures in Atg1016h mutant animals under the same growth condition as in B–B″. D, quantification of the number of RFP-Atg8a punctate spots from B′ and C′. 145 cells from 16 WT fat body samples and 114 cells from 15 Atg1016h mutant fat body samples were counted. E, quantification of the size of RFP-Atg8a punctate spots shown in B′ and C′. 1000 spots from 12 WT fat body samples and 198 spots from 16 Atg1016h mutant fat body samples were measured. F–F″, accumulation of GFP-Atg8a and RFP-Atg8a punctate structures in fat body cells of wandering-stage WT third instar larvae. G–G″, absence of GFP-Atg8a and RFP-Atg8a punctate structures in Atg1016h mutant animals. H, quantification of the number of RFP-Atg8a punctate spots shown in F′ and G′. 82 cells from 15 WT fat body samples and 162 cells from 29 Atg1016h mutant fat body samples. I, quantification of the size of RFP-Atg8a punctate spots shown in B′ and C′. 150 spots from six WT fat body samples and 410 spots from four Atg1016h mutant fat body samples were measured. Data are presented as mean ± S.E. An unpaired t test was used for statistical analysis. ***, p < 0.001. Scale bars: 50 μm.