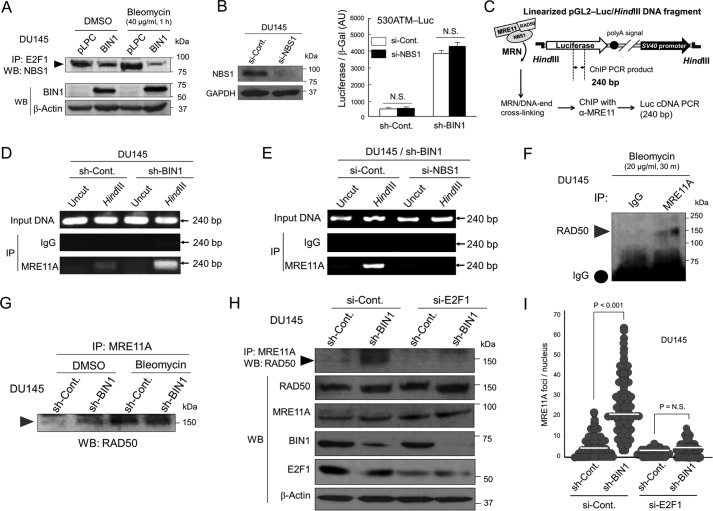
Figure 5.
E2F1 is vital for MRN formation in BIN1-deficient nuclei under optimal culture conditions. A, co-IP/Western blot analysis of endogenous E2F1/NBS1 protein complex in DU145±pLPC-BIN1 cell lines in the presence and absence of bleomycin (40 μg/ml, 1 h). B, NBS1 siRNA (si-NBS1) or scrambled control siRNA (si-Cont) was cotransfected with 530ATM-Luc in DU145±sh-BIN1 cells. The raw luciferase activities were normalized with the cotransfected β-gal (pcDNA3–β-gal: 1:10 (w/w)) activity. AU, arbitrary unit. N.S., not significant. C, schematic diagram of ChIP-based in vivo DNA end-binding assay. Cells were transfected with the pGL2-control luciferase (Luc) plasmid DNA linearized by HindIII restriction (pGL2/HindIII-Luc DNA fragment). The super-coiled (uncut) pGL2-Luc plasmid DNA was used as the negative control. D, to determine whether a BIN1 loss enhances the MRE11A/DNA end-binding activity, the indicated 240-bp region of the Luc cDNA was amplified by genomic PCR after an immunoprecipitation with an anti-MRE11A antibody in the DU145±sh-BIN1 cell lysates treated with formaldehyde. E, ChIP-based DNA end-binding assays verified that endogenous NBS1 is vital for the MRE11/DNA-end interaction. To deplete endogenous NBS1 protein, si-NBS1 was cotransfected. F, co-IP/Western blot analysis demonstrated the physical binding of endogenous MRE11A with RAD50 in the presence of bleomycin (20 μg/ml, 30 min). G, co-IP/Western blot analysis verified that similarly to bleomycin treatment, the loss of BIN1 stabilizes the MRE11A/RAD50 protein complex in vivo. H, co-IP/Western blot analysis revealed that the BIN1 loss-mediated stabilization of the MRE11A/RAD50 protein complex was canceled by depleting E2F1. I, scatter plot analysis of the MRE11A foci per nucleus in the DU145±sh-BIN1 (stable) cell lines after transient transfection of si-E2F1 or si-Control. The cells were counterstained with an anti-E2F1 antibody, and the number of MRE11A foci in si-E2F1-transfected nuclei was counted. Horizontal bars indicate mean values.