Figure 2.
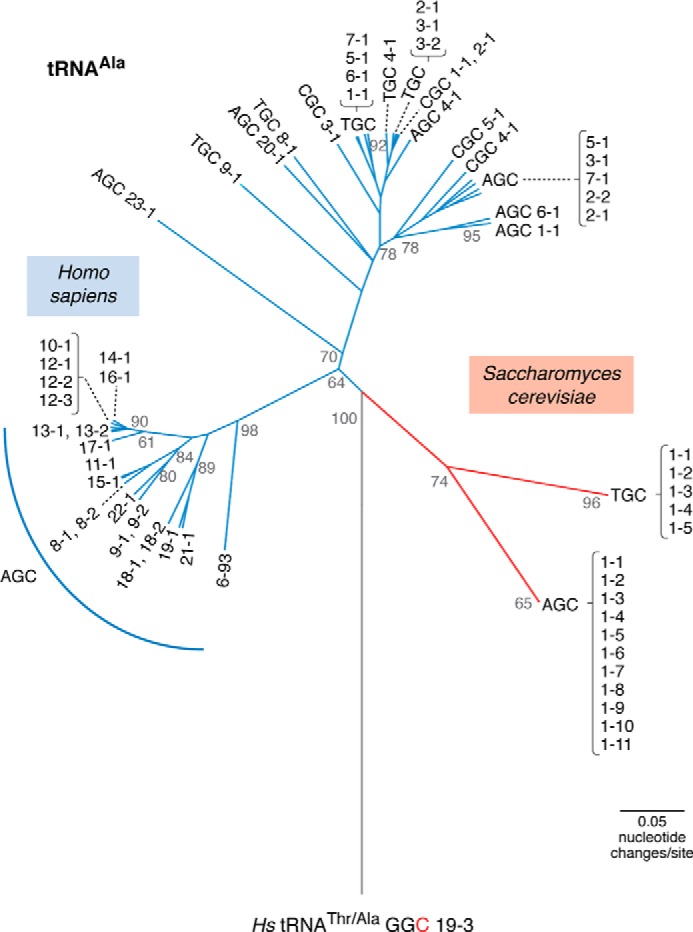
Phylogenetic relationships of human and yeast tRNAAla. The tree is based on an alignment of all known human and yeast tRNAAla iso-acceptors. The vastly expanded number and greater diversity of human tRNAAla genes compared with their yeast counterparts are evident. The tRNAs are labeled according to gene names in the genomic tRNA database (43), which include anticodon sequence followed by a numbering system where the first number indicates similar sequences, and the second number indicates a gene copy identifier. The human reference genome contains a misannotated tRNAAla, which was used to root the tree. This gene, tRNAAla-GGC-19-3, is a tRNAThr with a mutation (T36C) endowing the tRNA with an alanine anticodon. This is the only example of the alanine GGC anticodon in humans. Scale bar indicates the number of nucleotide changes per site in the tRNA sequences. The tree was calculated similarly as before (135). Briefly, a starting tree computed in MultiSeq 2.0 (136) was optimized to identify the maximum likelihood tree using PhyML 3.1 (137). Statistical branch support (out of 100) was calculated based on an approximate likelihood ratio test method (138) as implemented in PhyML.
