Abstract
Aminoacyl-tRNA synthetases (ARSs) are universal enzymes that catalyze the attachment of amino acids to the 3′ ends of their cognate tRNAs. The resulting aminoacylated tRNAs are escorted to the ribosome where they enter protein synthesis. By specifically matching amino acids to defined anticodon sequences in tRNAs, ARSs are essential to the physical interpretation of the genetic code. In addition to their canonical role in protein synthesis, ARSs are also involved in RNA splicing, transcriptional regulation, translation, and other aspects of cellular homeostasis. Likewise, aminoacylated tRNAs serve as amino acid donors for biosynthetic processes distinct from protein synthesis, including lipid modification and antibiotic biosynthesis. Thanks to the wealth of details on ARS structures and functions and the growing appreciation of their additional roles regulating cellular homeostasis, opportunities for the development of clinically useful ARS inhibitors are emerging to manage microbial and parasite infections. Exploitation of these opportunities has been stimulated by the discovery of new inhibitor frameworks, the use of semi-synthetic approaches combining chemistry and genome engineering, and more powerful techniques for identifying leads from the screening of large chemical libraries. Here, we review the inhibition of ARSs by small molecules, including the various families of natural products, as well as inhibitors developed by either rational design or high-throughput screening as antibiotics and anti-parasitic therapeutics.
Keywords: aminoacyl tRNA synthetase, antibiotic action, malaria, transfer RNA (tRNA), translation, inhibitor, aminoacyl sulfamate, mupirocin, peptidyl transferase, Trojan horse approach
Introduction
Protein synthesis in all living cells relies on the faithful decoding of mRNAs to produce polypeptide chains of the correct sequence. The genetic information encoded in the string of the trinucleotide codons of the message ultimately determines the amino acid sequence of the protein, governed by the specific pairing of aminoacyl-tRNAs and individual mRNA codons within the context of the translating ribosome. In addition to the accuracy of this codon–anticodon interaction in the decoding process, the overall fidelity of protein synthesis depends on formation of the aminoacyl-tRNA (1). This key reaction is catalyzed by a diverse family of enzymes, the aminoacyl-tRNA synthetases (ARSs),2 all of which synthesize aminoacyl-tRNA. Each ARS is specific for a particular amino acid and joins that amino acid to the tRNA that is specific for (or “cognate” to) the particular amino acid (2).
Given the essentiality of protein synthesis to the survival and fitness of living cells, it is not surprising that the translation apparatus represents one of the most frequently targeted cellular processes by natural product antibiotics. Because of their role as key agents in implementing the genetic code, ARSs are a ubiquitous and essential part of that apparatus. Their structures and functions have been characterized extensively in recent decades, and there is now a rich literature with detailed description of mechanistic features that are both common to the class as a whole and unique to specific families (3, 4). In addition to providing a foundation for synthetic biology engineering efforts, this information has also contributed to the development of novel and highly-potent inhibitors. Inhibitor development has also been stimulated by the growing discovery of noncanonical functions for ARSs, particularly in the areas of signaling, regulation, and overall maintenance of cellular homeostasis (5).
Natural antibiotics that target the peptidyl transferase and decoding centers of the ribosome, like chloramphenicol and streptomycin, respectively, provide examples of the potential clinical benefits of inhibiting the translational apparatus (6). By contrast, natural product inhibitors of tRNA synthetases were similarly identified decades ago, but with limited exceptions they have yet to gain general clinical acceptance. These natural products provide powerful validation for ARSs as therapeutic targets, but their history also illustrates some of the formidable barriers to their development into useful drugs. Recent advances in genomics, high-throughput screening of complex libraries, novel inhibitor frameworks, and the identification of new regulatory functions for ARSs have provided a new motivation to explore the potential clinical benefits of inhibiting select members of this class. Another important emerging dimension of research into ARSs concerns their involvement in diverse human diseases. For example, ARSs were identified decades ago as critical antigens in the autoimmune diseases of polymyositis and dermatomyositis (7, 8). Their specific roles in the pathophysiology of these diseases have yet to be fully explained. More recently, mutant versions of ARS genes have been identified in association with peripheral neuropathies, sensorineural disorders, and severe neurodevelopmental phenotypes (9). An improved understanding of small molecule modulation of ARSs may provide a new route to therapeutic interventions for these diseases.
This review provides an update on therapeutic inhibition of aminoacyl-tRNA synthetases, including the major classes of inhibitors, the most widely studied therapeutic targets, and perspectives on the most promising future opportunities. This topic has been addressed in previous reviews (10–15) that emphasized anti-ARS compounds as possible antibiotics and anti-infectives against both bacterial and eukaryotic pathogens. Following a brief introduction to the aminoacylation reaction, we will discuss efforts to target prokaryotic organisms, move on to eukaryotic pathogens, and then conclude with prospects for therapeutics against noninfectious diseases, including cancer and neurological diseases.
Aminoacylation reaction and the class structure of the aminoacyl-tRNA synthetase superfamily
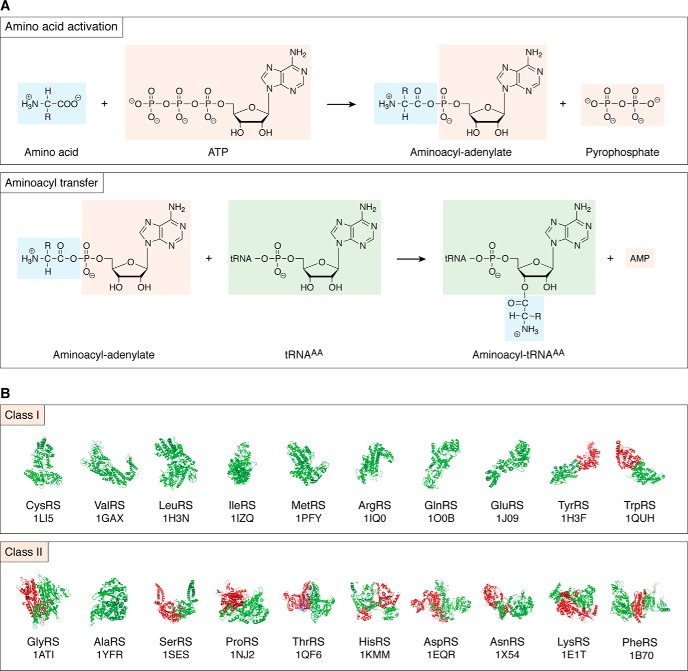
Prior to discussing specific inhibitors, it is useful to review the chemistry of the aminoacylation reaction and the fundamental class structure of the ARSs. The aminoacyl-tRNA synthesis reaction occurs in two distinct steps (16). In the first reaction, amino acid and ATP are condensed to form a mixed anhydride adenylate intermediate, with pyrophosphate serving as the second product (Fig. 1A). The adenylate intermediate is noncovalently bound to the active site. As we will see below, chemical derivatives of the adenylates are potent inhibitors of the corresponding ARSs and are an important starting point for further therapeutic developments. In the subsequent aminoacyl transfer step, the amino acid is esterified to the 3′ end of the tRNA in a regiospecific reaction. For some ARSs, there is evidence that the nonbridging oxygen of the α-phosphate of the AMP moiety participates as a general base to activate the tRNA (17, 18).
Figure 1.
Aminoacylation reaction and the two classes of aminoacyl-tRNA synthetases. A, two half-reactions of the aminoacylation reaction, showing structures of the reactants and products. For most tRNA synthetases, the amino acid activation reaction can occur in the absence of tRNA. In the case of glutamyl-tRNA synthetase (GluRS), glutaminyl-tRNA synthetase (GlnRS), and arginyl-tRNA synthetase (ArgRS), the presence of tRNA is required for amino acid activation. B, representative structures of each of the 20 standard ARSs, divided into the two principal structural classes. The RCSB Protein Data Bank ID numbers are indicated below each structure.
The ARSs divide into two classes of essentially 10 members each (a second LysRS represents an additional Class I enzyme) on the basis of the protein fold of the catalytic domain, characteristic signature sequences, and mechanistic features of the aminoacylation reaction (4, 19). Class I enzymes share a catalytic domain based on the Rossmann/nucleotide-binding fold, an α/β sheet with alternating α-helices and β-strands oriented in parallel fashion (Fig. 1B). Class I enzymes also possess HIGH and KMSKS signature sequences that constitute the ATP-binding site and provide interactions to maintain the ATP in an extended conformation. Class I enzymes typically approach the tRNA acceptor stem from the minor groove side. Finally, three of the Class I ARSs (ArgRS, GluRS, and GlnRS) require the presence of the tRNA for adenylate formation.
By contrast, the catalytic domains of Class II enzymes share a conserved seven-stranded antiparallel sheet and characteristic signature sequences that constitute the motifs 1–3. Motif 1 is an extended α-helix linked to a β-strand that contributes to the formation of the dimeric interface. Motif 2 (a conserved β-strand hairpin) and motif 3 (β-stand and α-helix) are core structural elements of the Class II catalytic domain antiparallel β-sheet that provide ATP recognition. In Class II ARS-active sites, ATP is in a “bent” conformation. Class I and II ARSs also differ with respect to the terminal hydroxyl of the 3′-terminal adenosine Ade-76 to which the amino acid is attached. Class I enzymes aminoacylate on the 2′-OH of the terminal ribose, whereas Class II enzymes typically aminoacylate on the 3′-OH. Class II ARSs also feature conserved acidic amino acids that coordinate Mg2+ ions that stabilize the pyrophosphate leaving group.
ARSs possess the ability to finely discriminate among different chemically similar amino acid substrates, employing both passive binding and active editing strategies (1, 20). For those ARSs that aminoacylate hydrophobic amino acids (e.g. IleRS, ValRS, LeuRS, ThrRS, and AlaRS), the amino acid pocket alone provides insufficient discrimination to prevent misacylation of near-cognate amino acids. The problem is particularly acute for pairs that differ by a single methyl or hydroxymethyl group (21). Thus, amino acid groupings consisting of Ile/Val, Leu/Ile/Met/Val, Val/Thr, Thr/Ser, and Ala/Gly/Ser have imposed selection for editing function in the ARSs associated with the first amino acid in the grouping. In the original “double sieve” model proposed to account for ARS discrimination, an initial “coarse” sieve prevents the binding and activation of larger and chemically dissimilar amino acids, whereas a second “finer” sieve allows amino acids smaller than the cognate to pass through to a second active site (22). These misacylated substrates undergo hydrolysis by a specific deacylation activity in the editing site. As predicted by this model, ARSs with well-defined editing properties possess separate protein domains dedicated to this editing function. This deacylation mechanism has been referred to as “post-transfer editing” (23).
Alternatively, the misactivated amino acid can be eliminated by hydrolytic decomposition of the adenylate prior to transfer of the amino acid to the tRNA, referred to as “pre-transfer editing” (24). The relative contribution of these two mechanisms depends on the relative rates of aminoacyl transfer and deacylation (25, 26). Evidence from both prokaryotic and eukaryotic systems indicates there is a fitness cost for the absence of editing functions (27–29). For example, an inbred mouse strain containing an editing-deficient AlaRS allele (the sticky (Sti) allele) exhibits Purkinje cell degeneration and associated deficits in cerebellar function (30). Some nonproteogenic amino acids are able to evade natural editing systems, with potentially toxic consequences (31, 32). Interestingly, editing functions may be diminished in certain obligate parasitic species, such as Mycoplasma (33, 34).
In Escherichia coli and other enteric bacteria, there is at least one ARS for each of the 20 amino acids. In other bacterial and most archaeal species, GlnRS and AsnRS orthologs may be absent, and indirect pathways employing amidotransferases are used to produce the corresponding aminoacylated tRNAs (35). Indirect pathways also account for the absence of CysRS in some Archaea genera (36). In humans, there are 38 nuclear-encoded ARS genes, apportioned equally among cytoplasmic and mitochondrial enzymes, but there are also two dual-compartment enzymes (LysRS and GlyRS) that function in both the cytoplasm and mitochondria (37). In human pathogens such as the apicoplexans, the number of ARS genes can vary, as some of the encoded enzymes function in both the cytoplasm and in a specialized organelle, the apicoplast (38). As a consequence of these multiple copies, an inhibitor directed against a pathogen that is specific for mammalian cells will typically face three or potentially four distinct but similar versions of an enzyme from a given ARS family. Accordingly, a particularly acute problem in the synthesis of any ARS inhibitor is achieving selectivity for the pathogen enzyme over the host cytoplasmic and mitochondrial ARS enzymes.
Targeting prokaryotic organisms: natural products and designed inhibitors
Pathogens challenged by new antibiotics engage multiple pathways that allow resistance to a drug to develop. These include acquiring mutations in the target gene of interest, acquisition of enzyme functionalities that can modify the drug itself, and development of strategies to minimize internal drug concentration, such as changes in permeability or efflux. All of these strategies present hurdles to the development of anti-ARS compounds as therapeutics.
During the period of 1990–2000, the potential of the aminoacyl-tRNA synthetases to serve as a new class of antibiotic targets was explored systematically (39). Potentially useful features of ARS that render them as attractive antibiotic targets include the following: (i) their essential function in all cells; (ii) a divergence between prokaryotic and eukaryotic versions that provides structural differences that can be exploited; (iii) strong conservation of gene sequences among bacterial species that suggests that prokaryotic-specific drugs might be of broad spectrum; and (iv) the presence of 20 different ARSs, all of which represent specific enzymes that can be targeted individually or in combination. Finally, the enzymes are soluble, readily purifiable, express well, and can be assayed in a high-throughput regime.
Structural analyses and sequence comparisons between ARSs provide detailed information about active-site differences that might potentially be exploitable for rational drug design (11, 13). Whereas ATP-binding determinants are conserved at the class level, active-site residues that recognize the amino acid substrate show significant conservation in the ARSs of individual families (i.e. all AlaRSs, for example) across multiple kingdoms. This suggests a strategy of inhibitor design in which common elements target the class-conserved ATP-binding pockets, and variable features target amino acid binding site residues.
Mupirocin: the natural product ARS inhibitor that validated ARSs as antibiotic targets
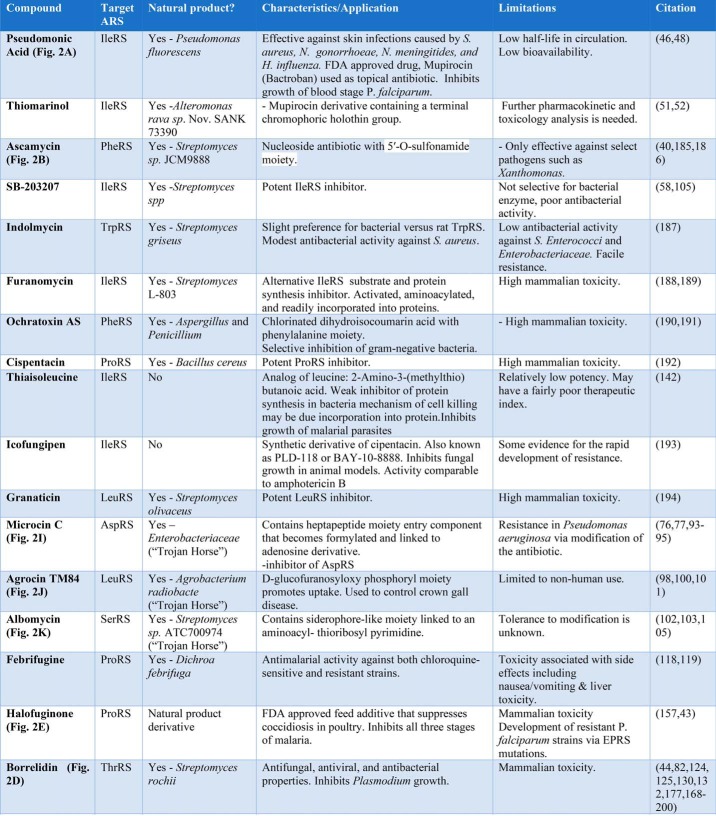
Natural products that inhibit the ARSs were first characterized several decades ago, and their chemical frameworks have served as important lead compounds in the development of antibiotics and antimalarials. Among the natural products, two are particularly noteworthy for their importance in the antibiotic realm. Pseudomonic acid (Fig. 2A) serves as the basis of the approved topical antibiotic mupirocin, and ascamycin (Fig. 2B) (40) represents a starting framework for the rational design of intermediate based inhibitors (IBIs). By contrast, several other natural products, including cladosporin (Fig. 2C) (41), borrelidin (Fig. 2D) (42), and halofuginone (a derivative of febrifugine) (Fig. 2E) (43), have shown significant promise as inhibitors of eukaryotic pathogens, including malaria. These are discussed in great detail in later sections.
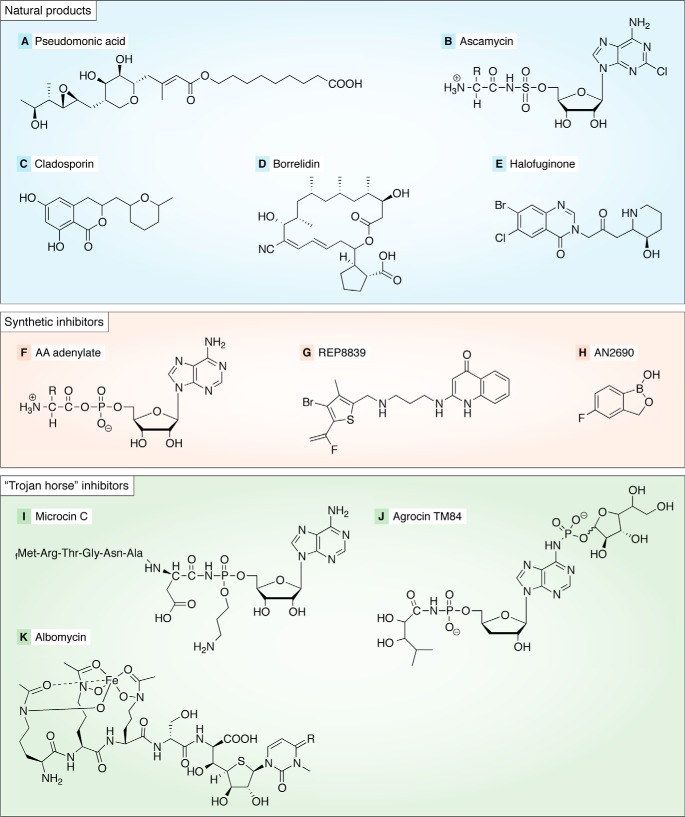
Figure 2.
Structures of representative natural and synthetic ARS inhibitors. A, pseudomonic acid parent structure; B, ascamycin (for ascamycin, R = −CH3; for dealanylascamycin, the alanyl moiety is absent altogether); C, cladosporin; D, borrelidin; E, halofuginone; F, generalized aminoacyl-adenylate, which is the basis of IBIs; G, REP8839; H, AN2690; I, microcin C; J, agrocin TM84; K, albomycin.
An important feature of many of the ARS natural product inhibitors is that their biosynthesis is accomplished by polyketide synthesis (PKS) protein complexes encoded by large gene clusters within the genomes of the Streptomyces or Pseudomonas species that most frequently produce ARS natural product inhibitors (44, 45). A typical indication of the ARS-specific nature of the inhibitors produced by the PKS modules of these clusters is that they encode a dedicated ARS gene that is distinct from the standard ARS ortholog and that confers immunity to the natural product inhibitor. The modular nature of inhibitor biosynthesis associated with these PKS clusters allows for the generation of relatively complex molecules with extensive modifications and high affinity for their ARS targets. A drawback is that the large number of asymmetric carbons and functional group decorations on these molecules provides a high barrier to rapid structure–activity relationship (SAR) investigations by standard organic chemistry. As will be discussed below, genome modification and semi-synthetic approaches have provided novel routes to new structural variants.
Because of its successful adaptation into an FDA-approved topical antibiotic (mupirocin), the natural product ARS inhibitor pseudomonic acid (PMA) represents one of the most important of all ARS inhibitors (Fig. 2A). PMA is a polyketide antibiotic produced by Pseudomonas fluorescens NCIBM 10586 (46) and comprises several different structures (MupA, -B, and -C) that differ slightly in both the polyketide and fatty acid portions (47). In all of these structures, the C17 monic acid moiety is linked to a C9 9-hydroxynonanoic acid fatty acid via an ester linkage through the hydroxy group. A 75-kb gene cluster in P. fluorescens encodes the PKS machinery that synthesizes PMA, including a mupirocin-resistant IleRS homolog that protects the producer cell from the effect of the antibiotic (48). A high cell density is an essential condition for mupirocin synthesis, and the regulon is under quorum-sensing control. PMA is highly specific for isoleucyl-tRNA synthetase, exhibiting on the order of 8000-fold selectivity for pathogenic IleRS over the mammalian enzymes (46).
Several X-ray structures of the IleRS–PMA complex have been determined (49, 50) and indicate that PMA binds to the cleft where aminoacyl adenylate formation occurs. The tetrahydropyran ring mimics the structure of the adenine ring and occupies the ATP-binding site, whereas the hydrophobic C12–C14, C17 terminus of the compound occupies what is normally the isoleucine pocket. In addition, the C-1′ to C-9′ 9-hydroxynonanoic moiety of PMA interacts with the loop corresponding to the Class I KMSKS signature sequence, maintaining it in a “closed” conformation that blocks the catalytic cycle (50). Some of these interactions are specific to bacterial IleRS, which accounts in part for the selectivity of PMA for prokaryotic targets. Thiomarinol is a variant of PMA that is produced by the marine bacterium Alteromonas rava sp. nov. SANK 73390 and contains a terminal chromophoric holothin group (51, 52). This may serve as a new scaffold for the development of further antibacterial PMA variants.
Although the relatively high affinity of PMA for IleRS and its selectivity for the prokaryotic enzymes are highly valuable properties, the labile ester bond in the structure provides for a relatively low half-life in circulation. Moreover, because of serum binding, PMA has poor bioavailability. Despite these limitations, PMA formulated as a 2% suspension in PEG is an FDA-approved topical antibiotic, mupirocin (commercial name Bactroban). Mupirocin is effective against skin infections caused by Staphylococcus aureus, Neisseria gonorrhoeae, Neisseria meningitidis, and Haemophilus influenzae. Repeated use of mupirocin in the clinic typically leads to the selection of resistant microorganisms via two distinct routes (53). The first is low-level resistance, which occurs via selection for drug-resistant versions of the resident chromosomally encoded IleRS. The higher-level resistance is mediated by a plasmid shared by resistant bacteria that encode for a eukaryotic homolog of IleRS that is refractory to high levels of mupirocin. Whereas mupirocin represents a significant “proof of principle” compound for anti-ARS therapeutics, it suffers from the facile development of resistance when applied repeatedly, a concern that is common to other potential ARS-directed antibiotics (54).
The success of the parent compound has inspired synthetic efforts to develop versions of mupirocin with a higher affinity for the parent IleRS. One strategy involves the generation of hybrids between pseudomonic acid and derivatives of the Ile-AMP adenylate. Some of these rationally designed variants of mupirocin (55–57) exhibit extremely high affinity for IleRS (Ki <0.001 nm) (56). In addition to PMA, other natural product IleRS inhibitors based on chemical frameworks distinct from PMA include SB-203207, which was isolated from Streptomyces spp, (58, 59). These derivatives showed nanomolar inhibition but low selectivity for bacterial species and low antibacterial activity.
In addition to PMA and the aforementioned natural products that target eukaryotic pathogens, there are natural product ARS inhibitors whose potency/toxicity profiles have discouraged more extensive investigation. Among the examples of this class included in Table 1 are indolmycin, which targets TrpRS; ochratoxin, which targets PheRS; and cis-pentacin, which targets ProRS. By and large, these molecules are reasonably potent inhibitors of both pathogenic prokaryotic and host eukaryotic enzymes, but the specificity for the former is too low to merit deeper investigation. Despite the relatively simple chemical structures of some of these compounds, none have been investigated by SAR chemistry to see whether the toxicity to host cells can be attenuated.
Table 1.
Natural product and synthetic inhibitors of aminoacyl-tRNA synthetases
Identification of antibiotic leads derived from ARS inhibitors by use of rational design and high-throughput screening
The significant challenges associated with synthesizing variants of the aforementioned natural products have limited their exploitation as potential therapeutics. As a result, much of the prior efforts by pharmaceutical and biotech companies to therapeutically target the ARSs have relied on two basic approaches: (i) rational design approaches based on bona fide aminoacylation reaction intermediates, and (ii) high-throughput screening (HTS) of unbiased chemical libraries of high complexity. Although both strategies have been used successfully to identify inhibitors of prokaryotic ARSs with relatively small inhibition constants (Ki <50 nm), neither has yielded an antibiotic approved for use in the clinic. As will be seen in a later section, an unbiased phenotypic screening approach was ultimately successful in the generation of a clinically useful therapy for a eukaryotic pathogen.
Many of the rational design strategies begin with the aminoacyl adenylate (Fig. 2F), an obligatory intermediate in the aminoacylation reaction catalyzed by all ARSs, irrespective of class. There are two inherent problems with a specific aminoacyl-adenylate for a particular ARS as a potential lead for therapeutic inhibitors. First, the mixed anhydride acylphosphate bond of the intermediate is readily susceptible to hydrolysis. Second, the charged and polar nature of the intermediate will preclude passage across cell membranes without the intervention of a specific transporter. By replacing the phosphate with nonhydrolysable “biosteres” that retain the tetrahedral geometry of the phosphate, the potential hydrolysis of the acylphosphate group is eliminated (39). Key functional groups that interact with the enzyme can be retained. Chemical linkers can be chosen to recapitulate the negative electron density around the acylphosphate, as well as preserve the molecular dimensions of the adenylate.
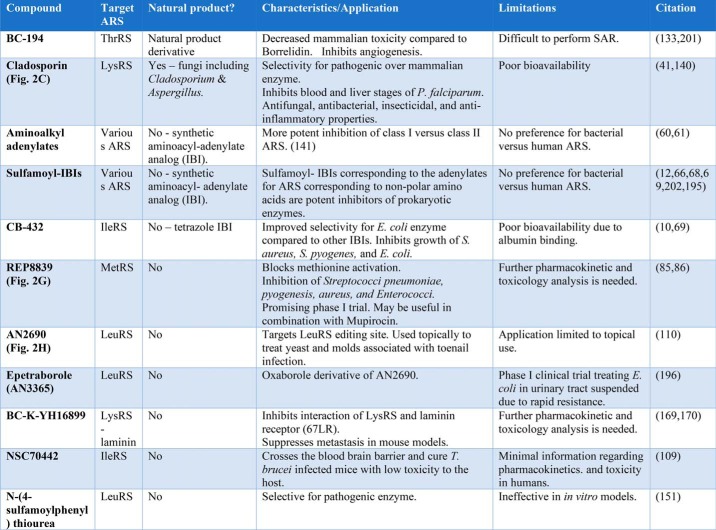
The amino acid alkyl adenylates and the aminoacyl sulfamates represent two classes of IBIs that have been investigated systematically. IBIs bind to their active sites of their respective ARSs with affinities in the high picomolar to low nanomolar range. These dissociation constants are 2–3 orders smaller than that of the natural substrates ATP and amino acid (10). Amino alkyl adenylates leave the phosphate intact but substitute the carbonyl with a methylene group to minimize hydrolysis (60, 61). The inhibition constants for amino alkyl adenylates range from a Ki(Met) of 4.7 nm for E. coli MetRS to a 60% inhibition at 300 mm for S. aureus ThrRS (61). Overall, the amino alkyl adenylates were found to be much better inhibitors of Class I enzymes than Class II enzymes, likely because the carbonyl oxygen makes a critical interaction in the Class II ARS active site (61).
The sulfamoyl-based IBIs in which the anhydride linkage is replaced by a 5′-O-sulfonamide moiety are similarly potent inhibitors. These are also referred to as aminoacyl sulfamoyl adenosine derivatives, or as AA-AMS with the AA replaced by the corresponding amino acid. As seen with amino alkyl adenylates, they exhibit virtually no preference for bacterial over human enzymes. Of note, the sulfamoyl analogs are chemically synthesized versions of the natural product ascamycin (Fig. 2B), but lack the chlorine atom. Aminoacyl adenylate sulfamoyl analogs have been valuable co-crystallization aids with ARSs and useful proxies for the adenylate in investigations of its effect on tRNA recognition (62–65). In general, the sulfamoyl-IBIs corresponding to the adenylates for ARS charging nonpolar amino acids (including isoleucyl-, leucyl-, and valyl-tRNA synthetase) are relatively potent inhibitors of pathogenic prokaryotic enzymes (i.e. from S. aureus) with IC50 values in the range of 2–30 nm (66). Although less work has been done to characterize the sulfamoyl adenylates based on more polar amino acids, the Glu-AMS analog proved to be a more potent inhibitor for the respective E. coli GluRS then Gln-AMS for its cognate enzyme. These differences were interpreted to suggest that Glu-AMS is a better transition state analog for its cognate enzyme than Gln-AMS for GlnRS (67). Because of their poor uptake by cells, comparatively little work has been done investigating the in vivo use of these compounds (68).
As noted above, a major obstacle to clinical use of the sulfamoyl adenylate-based IBIs is their relatively poor selectivity for prokaryotic pathogen ARSs versus the ARSs of the host. To obtain better selectivity for prokaryotic enzymes, the adenine in IBIs has been substituted by tetrazoles and other heterocycles, producing better selectivity for the E. coli enzyme, but not increasing overall potency. One successful result of this strategy was the compound CB-432, which exhibited 60–1100-fold discrimination for the pathogenic IleRS enzyme (10). CB-432 inhibited growth of S. aureus, Streptococcus pyogenes, and E. coli in culture with MICs in a range of 0.5–10 μg/ml. Because of poor bioavailability, limited success was achieved in mouse models, with some 98.5% of the drug remaining bound to albumin (69). Although this example highlights the potential utility of modifying the IBI framework, problems with bacterial uptake again proved to be a barrier limiting the utility of these compounds as actual drugs (70, 71).
More recently, the IBI approach has been augmented by exploring modifications to the basic chemical framework, employing in silico modeling against ARS active sites as a screening tool to tailor functional group changes. Recent examples include inhibitors of ThrRS and LeuRS, which featured a benzene sulfonamide core and extra substituents that confer specificity for the ARS amino acid–binding pockets (72, 73). In the case of the compounds targeting LeuRS, the benzene sulfonamide scaffold was improved by docking simulations employing the E. coli LeuRS structure and by the use of isothermal titration calorimetry to assess binding affinity. By adding amino pyridine or amino pyrimidine substituents to the meta position of the benzene sulfonamide, leads could be obtained with low nanomolar affinity for E. coli LeuRS and micromolar affinity to human LeuRS. Of interest, adding lipophilic substituents improved the binding profile, such that enthalpic contributions to binding predominate. For the final compounds, the best MICs were obtained with E. coli among representative Gram-negative organisms; there was virtually no inhibition of S. aureus.
Other modifications of the IBI scaffold have been tested to limit hydrolysis of the acylphosphate group in the adenylate, which might improve stability and bio-availability. The most interesting of these involves substituting the adenosine moiety with 3-deazaadenosine, thereby blocking a potential side reaction involving the base N3 group. The side reaction leads to hydrolysis of the sulfamoyl moiety, and formation of N3,C5′-cycloadenosine (74). Although the resulting derivatives (abbreviated here as aaS3DAs) of Class II ARS adenylates demonstrated very little inhibitory activity, excellent potency was achieved with the leucyl version, essentially equivalent to the parent Leu-AMS. Interestingly, the lack of inhibition of Class II ARSs by aaS3DAs could be due to the stabilizing interactions between the N3 of adenine and a conserved carboxylate and polar backbone functionality in motif 3. Whereas Class I-based aaS3DA versions initially appeared more promising, the leucine aaS3DA failed to inhibit growth of any of the members of a panel of Gram-positive and Gram-negative bacteria, essentially replicating prior observations with unmodified AA-AMSs (71, 74–77).
Despite the failure to obtain a “breakthrough” lead compound by this approach, an important conclusion from these studies is that use of advanced modeling and theory tools such as quantum mechanical chemical calculations can provide additional insights into the interactions of these compounds with ARS active sites, thereby rationalizing the significant differences in Class I versus Class II ARS-targeted compounds (74). Such approaches extend the insights from existing ARS–inhibitor complexes, whose numbers are growing in the RCSB Protein Database. Many ARS complexes with aminoacylation active-site inhibitors have been reported, particularly those of IleRS, TyrRS, TrpRS, ProRS, LysRS, and ThrRS with their specific molecules (72, 78, 79). Structures are also available for the IleRS–PMA complex (49, 50). A common theme for these inhibitors is that the sites of inhibitor binding overlap with binding sites for the amino acid, ATP, or both (Fig. 3C). The only case where the binding site clearly overlaps with a tRNA-specific subsite is in the case of borrelidin (Fig. 3C). Interestingly, binding of pseudomonic acid and borrelidin to their respective active sites prevents full cleft closure seen with the AA-AMS derivatives (56, 80–82).
Figure 3.
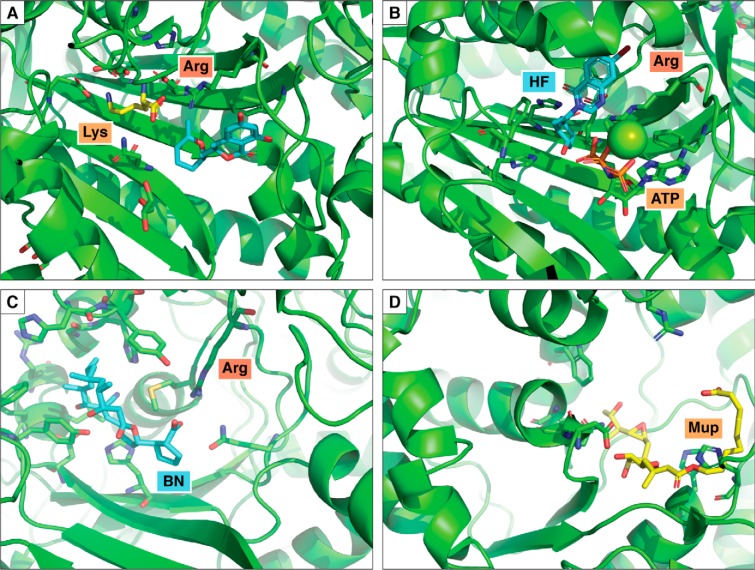
X-ray structures of selected ARS–inhibitor complexes. In each case, the secondary structure of polypeptide chain is depicted in ribbon representation (green), and the inhibitors are depicted in stick representation (blue or yellow). Selected active-site residues are shown in each panel, including the conserved motif 2 arginine. A, complex of P. falciparum LysRS with lysine and cladosporin (cyan) (PDB code 4YDQ). B, complex of HF with Homo sapiens ProRS (PDB code 4HVC). C, complex of H. sapiens ThrRS and borrelidin (PDB code 4P3N); D, complex of E. coli IleRS with mupirocin (PDB code 1JZS).
As an alternative to the rational design of ARS inhibitors using chemical scaffolds derived from IBIs, complex chemical libraries can be screened to identify hits based on their ability to inhibit the aminoacylation reaction, as measured using scintillation proximity assay technology. In one approach, a chemical compound collection of known structures was screened to identify features reminiscent of an AA-AMP adenylate (83). Using E. coli MetRS as a target, hits of medium potency (IC50 = 237 nm) were obtained. Alternatively, completely unbiased approaches have been used. Screening of a modest complexity chemical library (∼50,000 compounds) against 17 synthetases from S. aureus and Enterococcus faecalis produced hits for 5 of 17 ARSs that were derived from more than two distinct chemical structural classes of inhibitors. Whereas more hits were obtained for those ARSs that aminoacylate hydrophobic amino acids, such as PheRS, the IC50 values of these compounds were still relatively modest, in the low micromolar range (11).
Perhaps the greatest impact of HTS screening in the ARS domain has been to identify new inhibitor frameworks that might not have been predictable on the basis of prior structure knowledge. For example, quinoline carboxylic derivatives form the basis of several leads that have been pursued in detail, including one that passed into clinical trials. Among the first quinoline carboxylic inhibitors was a compound that targets the Candida albicans ProRS with high affinity (IC50 ∼ 5 nm) and good selectivity for the pathogen enzyme relative to the human enzymes (84). Soon thereafter, the important quinolone derivative REP8839 was developed by modification of an earlier fluorovinylthiophene compound identified from an HTS program targeting MetRS (85). REP8839 (Fig. 2G) blocks the methionine activation reaction by a mechanism uncompetitive with ATP and showed promising action against Gram-positive S. aureus, Streptococcus pneumoniae, S. pyogenes, and Enterococcus in phase 1 clinical trials. Activity was substantially lower on human cytoplasmic and mitochondrial MetRS (86). An alternative dual strategy that was tested featured the combined use of mupirocin and REP8839 as a topical antibacterial (39). This approach showed promise as a means to limit the emergence of resistant pathogens (see below) (87).
As of this writing, major pharmaceutical concerns appear to have largely suspended the search for new antibiotics targeting the ARSs, creating opportunities for the academic sector and earlier stage companies to continue this approach. Such efforts benefit from the accumulation of ARS structural and mechanistic information and the highly robust screening and new assay technologies that are available (88). Among the most promising work in this area is the effort to target a variety of ARSs from Pseudomonas aeruginosa, including HisRS, GluRS, and PheRS. Inhibitors with IC50 values in the 2–20 μm range have been identified and exhibit a bacteriostatic mode of inhibition when used to challenge a panel of pathogenic bacteria (89–92). These results suggest that only a relatively small portion of ARS inhibitor chemical space has been identified and that more potent compounds await discovery.
Trojan horse natural product antibiotics: a novel solution to the cell-barrier problem
As noted above, a major chief limitation of IBIs is their poor uptake by bacterial cells. An important new class of natural products that address this problem are the so-called “Trojan Horse” inhibitors. The name alludes to a two-part structure consisting of an uptake moiety that is readily cleaved off upon entry into the cell (the “Trojan Horse”) and a nucleotide toxin moiety (the “warhead”) that is a stable chemical analog of the AA-AMP intermediate. Microcin C (Fig. 2I) is a well-characterized representative of this antibiotic family produced in the Enterobacteriaceae that feature a post-translationally added heptapeptide moiety as the entry component and an aspartyl-AMP derivative as the warhead (93, 94). The peptide is modified by a formyl group at the N terminus and links to an adenosine derivative via a phosphoramidate bond. In contrast to the standard aminoacyl adenylate, the phosphate is modified by a 3-aminopropyl group. This latter group binds to a pocket adjacent to the aspartyl site. The transport of microcin C is facilitated by the permease YejABEF transporter. Once inside the cell, the peptide is processed by the processive digestion and peptidase action of PepA, PepB, and PepN and other enzymes, including deformylase and peptidases. These release a stable adenylate analog inhibitor that competes with aspartate for binding to AspRS, with the 3-aminopropyl group binding to a pocket adjacent to the aspartyl site (95). Only the processed form is inhibitory. Microcins are bacteriostatic inhibitors of Gram-negative bacteria, including E. coli, Klebsiella, Salmonella, Shigella, and Proteus (14). Of interest, the resistance of P. aeruginosa strains against this antibiotic arises from the activities of MccE and MccF, which catalyze acetylating and peptide hydrolase functions, respectively (96, 97). Both serve to provide resistance by modifying the antibiotic.
Agrocin TM84 (Fig. 2J) is a Trojan Horse inhibitor of LeuRS and targets Agrobacterium radiobacter K84 (98). Agrocin is a 9-(3′-β-d-2,3,)threopentafurano adenine nucleoside phosphoramidate conjugate. The targeting group is a d-glucofuranosyloxy phosphoryl moiety (99) that helps the antibiotic mimic the uptake of agrocinopine through the opine transporter by a susceptible host. (Opines are low-molecular-weight nitrogen-rich compounds produced by parasitic bacteria of the genus Agrobacterium. They accompany infection by the bacteria, which produces plant crown gall tumors. The opines are produced biosynthetically by the bacteria and represent a source of nitrogen and energy.) The adenylate moiety, a potent LeuRS inhibitor, is referred to as TM84 and features a stable 5-N-acylphosphoramidite bond rather than the standard phosphoanhydride. Interestingly, this molecule does not inhibit the activation reaction and requires the participation of the tRNA for inhibition (100). The agrocin 84–producing strain carries a protective version of LeuRS (AgnB2) (98, 101) that is analogous to the corresponding ARSs encoded in the Pseudomonas and Streptomyces genomes that carry protective alleles providing immunity against pseudomonic acid and borrelidin, respectively.
The last example of a Trojan Horse anti-ARS inhibitor is albomycin (Fig. 2K), which is produced by a complex pathway in Streptomyces sp. ATC700974 (102). The structure of albomycin consists of an aminoacyl-thioribosyl pyrimidine toxin that is linked to a siderophore-like moiety that promotes cellular uptake. Some variations on the basic structure are known (103). The siderophore moiety confers transport via a ferrichrome ABC transport system. After transport, peptidase N cleaves the peptide bond of albomycin, releasing the toxin. The resulting moiety, SB-217452, inhibits SerRS with a Ki in the range of 8 nm (59, 104, 105). This framework has recently proven amenable to modification by medicinal chemistry, opening the door to the synthesis of other derivatives (106). As seen with other ARS inhibitor natural product–producing strains, a special SerRS allele provides immunity against the toxin.
Because of their apparently unfettered ability to promote transfer of high-affinity ARS inhibitors (the “warheads”) into cells, these Trojan Horse antibiotics represent an especially promising avenue for future ARS therapeutic development. Although further work will be necessary to define the best approaches to achieve site-specific modification and to define their cellular tropism, the high potency already exhibited underscores the potential validity of these anti-ARS inhibitors.
Targeting eukaryotic pathogens with ARS inhibitors
Eukaryotic parasites exert a heavy disease burden on mankind worldwide, particularly in less developed countries. Among the most important pathogens worldwide are the Apicoplexans (including Plasmodium, Toxoplasma gondii, and Cryptosporidium) and the nematodes/platyhelminths, which collectively infect nearly a billion people. A representative disease of the latter is lymphatic filariasis, which is caused by the tropical nematodes Wuchereria bancrofti, Brugia malayi, and Brugia timori. No vaccines are available for the majority of these diseases. Antiparasitic drugs are compromised by the development of widespread drug resistance (107, 108). Significantly, ARS inhibitors show promise against the eukaryotic parasites that are the causative agents of a number of these diseases, representing a critical new area of research.
All of the pathogens referenced above are intracellular obligate parasites, with tropism for specific cell types that include erythrocytes, epithelial cells of the intestine, and other tissues. The dependence of these pathogens on translation function highlights the ARSs as plausible anti-parasitic targets. However, serious challenges include the extent of sequence conservation between the ARSs of a eukaryotic pathogen and a eukaryotic host, and the fact that some of the pathogens pass through relatively quiescent life stages that are difficult to address with inhibitors that attack protein synthesis. Despite these challenges, the presence of specialized organelles (like the apicoplast of the apicoplexans) in some of these pathogens that are absent in mammalian cells does present opportunities for therapeutic targeting.
The ARSs of eukaryotic pathogens possess multiple potential target sites, including 1) the synthetic/catalytic domain–binding pocket; 2) the editing domain, if present; 3) anticodon/tRNA–binding area; 4) allosteric binding sites; and 5) parasite-specific domains (109). Potential eukaryotic pathogens suitable for therapeutic targeting by ARS inhibitors range from relatively simple fungal infections to complex tropical diseases like malaria that involve multistage life cycles. The first ARS inhibitor specifically targeting a eukaryotic enzyme to gain success in the clinic is the fluorinated benzoxaborole AN2690 (also known as tavaborole; trade name Kerydin) (Fig. 2H). This LeuRS inhibitor was first identified in a yeast phenotypic screen of a library of boron-containing compounds, and its mechanism of action features the formation of a covalent bond between the boron and the diol of the tRNA's Ade-76 ribose in the LeuRS-editing active site (110). AN2690 is the first example of an anti-ARS drug that targets the editing site of an ARS, and it remains the best “proof of principle” of the therapeutic potential of the ARSs as targets for eukaryotic pathogens. When applied topically, the drug readily penetrates and effectively suppresses the growth of yeast and molds associated with onychomycosis (toenail infection).
Oxaboroles have also shown promise as agents against eukaryotic pathogens, e.g. African trypanosomiasis (111). Efforts to expand the application of oxaboroles to prokaryotic pathogens have had mixed results. A phase I clinical trial with epetraborole (GSK2251052/AN3365) against E. coli responsible for urinary tract infections had to be suspended because of the rapid emergence of resistant bacteria (112). More recently, a library of oxaboroles was screened against Mycobacterium tuberculosis, producing hits with MICs in the range of 1–2 μg/ml and IC50 values against LeuRS in the range of 0.64–3.5 μm (113). Although the best hits did produce decreases in lung and spleen cfu in a mouse TB model, the decreases were not as great as those achieved with the first line drug isonicotinylhydrazide. These promising results indicate the possible clinical application of oxaboroles against TB, perhaps as part of a potential combination therapy incorporating current frontline drugs. AN2690 and all its derivatives are highly specific for leucyl-tRNA synthetases, and efforts to extend the compounds to other tRNA synthetases have not been successful.
Despite the difficulties in trying to apply the benzoxyborole inhibitor framework to other ARSs, we consider AN2690 to be a major milestone in ARS inhibitor development, both as the first successful antifungal agent, and as a proof of principle example of the potential value of specifically targeting an ARS-editing site.
Malaria as a target of anti-ARS compounds
A large number of natural products target various ARSs in Plasmodium falciparum (the causative agent of malaria), offering promise that this pathogen may well be the next eukaryotic target that yields a successful anti-ARS therapeutic (114, 115). Malaria proceeds through multiple stages during its development, starting first with an insect form (the sporozoites), which upon transmission attacks liver cells (116). Following maturation, the sporozoites mature into schizonts that are released from the cells as merozoites. This form invades red blood cells that multiply asexually to form ring stage trophozoites. These can cycle as schizonts or release gametocytes. A total of 37 different potentially targetable ARS genes have been identified in Plasmodium, including 14 in the cytoplasm, ∼20 in the apicoplast, and several others that are localized to multiple compartments (38). The dual localization of AlaRS, GlyRS, CysRS, and ThrRS in the cytoplasm and apicoplast renders them targets of potential value (117). Accordingly, there are many ARS targets in Plasmodium, and multiple stages that may be vulnerable to therapeutic interdiction.
Natural product inhibitors of three different cytoplasmic ARS enzymes (GluProRS, ThrRS, and LysRS) have been explored as leads for antimalarial therapeutics. Febrifugine, a plant alkaloid from Dichroa febrifuga Lous., has long been part of Chinese pharmacopoeia, and extracts from D. febrifuga provide antimalarial activity against both chloroquine-sensitive and chloroquine-resistant strains (118, 119). Because of the harsh side effects associated with febrifugine, programs to synthesize less toxic variants led to the development of halofuginone (7-bromo-6-chloro-3-[-3-(3-hydroxy-2-piperdinyl-)-2-oxopropyl]-4(3H)-quinazolinone, abbreviated as HF) (Fig. 2E). In addition to its antimalarial properties, HF is an FDA-approved feed additive to suppress coccidiosis in poultry production (120). HF binds with high specificity to the prolyl-tRNA synthetase active site and is a potent inhibitor of proline incorporation in protein synthesis (43). The interaction of HF with glutamyl-prolyl-tRNA synthetase (EPRS) requires the presence of ATP, which stabilizes the local conformation of the active site to promote drug binding (121). The various moieties of HF mimic chemical features of the proline substrate and the 3′ end of the tRNA.
HF is active on all three stages of malaria (122), but HF-resistant strains of P. falciparum can arise via mutations in the portion of the EPRS gene encoding ProRS (123). Consistent with the binding of HF to the proline-binding site of ProRS, the addition of excess proline to culture media reduces the sensitivity of malarial parasites to HF. The use of HF in the clinic is limited more by the toxicity of the compound to animal cells than by the efficacy of killing of the malarial parasites themselves. By generating a secondary alcohol at the position in HF that is normally a ketone, a new lead (2′S,2R,3S)-halofuginol) has been generated with 65-fold better selectivity for P. falciparum, while still retaining excellent potency in the standard murine Plasmodium berghei ANKA strain liver stage model. HF also elicits a variety of stress responses in animal cells linked to amino acid starvation that may ultimately prove to be useful for other clinical indications, including autoimmunity, fibrosis-related diseases, and cancer (described in more detail below).
Another important anti-ARS natural product antimalarial is borrelidin (BN) (Fig. 2D), a C18 polyketide macrolide antibiotic produced in Streptomyces rochei. BN was originally discovered as a broad-spectrum inhibitor of viruses and bacteria (124, 125). More recently, BN has been shown to be a potent antifungal (126, 127). BN was also independently identified in a screen of natural antimalarials (128). Despite these promising activities, BN exhibits toxicity to animal cells, providing a deterrent to clinical exploitation.
The most important target of BN is ThrRS, although there has been debate about whether or not interactions with non-ThrRS targets in part explain the action of BN on animal cells (129). BN exhibits slow tight-binding kinetics with ThrRS in vitro with a Ki in the low nanomolar range (129, 130), and its binding site constitutes a relatively large portion of the ThrRS active site that overlaps with the amino acid and tRNA acceptor end–binding sites (Fig. 3C) (82). The competition between BN and threonine for the ThrRS active site accounts for the induction of the amino acid starvation response by BN, which can lead to cell cycle arrest and apoptosis above a critical BN concentration threshold that depends on cell type (130, 131).
BN is a strong inhibitor of Plasmodium growth in culture, targeting the cytoplasmic Plasmodium ThrRS with high potency and producing immediate inhibition of parasite growth (128). Pilot experiments in which mice infected with malaria were treated with borrelidin also produced promising results (132). To mitigate the toxicity of BN to animal cells, a novel strategy (“mutasynthesis”) combining rational engineering of Streptomyces strains and culturing cells with primer compounds has been used to produce derivatives of borrelidin that exhibit reduced toxicity with the human enzymes yet still potently retain other BN properties (133). When compared against a set of known ARS inhibitors (including mupirocin, cispentacin, and benzoxaboroles), a number of these BN derivatives exhibited very high potency (0.97 nm) against in vitro cultures of P. falciparum (134–136). New versions of BN have been isolated from marine organisms (137, 138), and new methylation variants have been developed by genetic modification of individual PKS modules in S. rochei producer strain (139). In addition to inhibition of malaria, BN exerts other physiological effects (described in later section) that are therapeutically significant.
The third ARS directed natural product is cladosporin (CS), originally identified in an unbiased screen of natural products that showed highly potent (∼50 nm) inhibition of blood and liver stages of P. falciparum (140). The structure features a 2,6-disubstituted tetrahydropyran fused to an isocoumarin derivative (Fig. 2C). CS inhibits blood and liver stage P. falciparum at nanomolar concentrations (140). Cladosporin is produced by fungal genera that include Cladosporium, Aspergillus, and others, and it had been previously shown to have a wide range of antimicrobial, insecticidal, and counter-inflammatory effects (41). The biosynthesis is accomplished via the action of a reducing/nonreducing iterative type I polyketide synthase complex encoded by a large biosynthetic gene cluster (45). As with other natural products, the cluster encodes a paralog of LysRS whose active-site substitution pattern is consistent with immunity to cladosporin. (The two other cladosporin LysRS orthologs do not contain such substitutions.) Based on the results of in silico docking experiments, cladosporin binds to the ATP-binding site of the cytoplasmic P. falciparum LysRS (141). Increasing concentrations of lysine do not affect the inhibition properties. Whereas the high selectivity of CS for the pathogen enzyme over the mammalian enzyme highlights its potential as an anti-malarial, its therapeutic exploitation has been limited by poor bioavailability. Efforts are also underway to target the apicoplast LysRS by use of synthetic compounds that mimic the LysRS adenylate.
The final ARS inhibitor natural product framework with potential against malaria is PMA (mupirocin), which inhibits growth of blood stage P. falciparum at nanomolar concentrations (142). This effect has been attributed to targeting of the apicoplast function. Unfortunately, mupirocin provides only minimal protection to P. berghei–infected mice, because of its low bioavailability. Given that compounds that induce amino acid starvation response for isoleucine (the apicoplast IleRS is a particularly sensitive ARS in this regard (143)) are potent antimalarials, other inhibitors that could potentially be explored as antimalarials include thiaisoleucine and icofungipen (Table 1).
Inspired by the promising results with natural product inhibitors, other recent efforts have used synthetic organic chemistry to identify new antimalarials from ARS inhibitors. The single AlaRS gene in Plasmodium is a dual-function enzyme that is essential for both cytosolic and apicoplast translation. By use of homology modeling to derive a Plasmodium AlaRS structure and docking simulations, a lead compound (4-(2-nitro-1-propenyl)-1,2-benzenediol) was obtained that could inhibit parasite growth with minimal effect on the mammalian enzyme (117). In addition, a recent high-throughput screening effort led to the identification of a series of bicyclic azetidines that target the cytoplasmic P. falciparum PheRS (144). Proliferation of malaria in the presence of these compounds is effectively blocked after only a single dose, and multiple stages of the parasite life cycle appear to be inhibited. Given the availability of a robust screening assay and the ability to perform SAR, these compounds should provide an excellent platform on which to develop improved therapeutics.
ARS inhibitors of nematode enzymes
In addition to malaria, another major class of eukaryotic pathogens with potentially targetable ARSs are the nematodes. Significant human pathogen infections include ascariasis, trichuriasis, hookworm, enterobiasis, strongyloidiasis, filariasis, trichinosis, and angiostrongyliasis (rat lungworm disease). Because of its role as a major immunodominant antigen, AsnRS from B. malayi has been investigated as a promising target in the search for therapeutics against lymphatic filariasis (145, 146). By HTS and docking simulations, a number of sub-micromolar inhibitors have been identified (147). These can potentially be further optimized by incorporating new structural data. Other inhibitors of the B. malayi AsnRS have been identified based on a pre-transfer editing assay (148). Some of these compounds potently inhibit the B. malayi AsnRS enzyme and kill adult B. malayi. A second important nematode disease is African trypanosomiasis, for which the Trypanosoma brucei is the pathogen responsible. For this pathogen, a number of ARSs show promise as targets (149). The ileRS gene is essential for T. brucei growth, and inhibitors that resemble the Ile-AMP intermediate (e.g. NSC70442, reported in Ref. 109) are able to cross the blood–brain barrier and cure T. brucei–infected mice with low toxicity to the host (150). LeuRS from T. brucei represents another major nematode target, and homology modeling in silico screening programs produced a few modest affinity hits (151). The best compound, an N-(4-sulfamoylphenyl)thiourea, could be predicted to make multiple hydrophobic and hydrogen bond interactions with the active site and produce a respectable IC50 of 1.1 μm. Despite good selectivity for the pathogen enzyme, these compounds have shown little effect in culture. Benzoxaboroles structurally related to AN2690 have been employed as lead compounds to target LeuRS from T. brucei, with IC50 values in the range of 1.6 μm (152). More recently, some of these same compounds have been tested against Cryptosporidium and Toxoplasma (153).
Targeting noninfectious diseases with tRNA synthetase inhibitors
The relationship between modulation of protein synthesis and modulation of the immune system is complex. The expansion of clonal B cell populations is a key part of the adaptive immune response, and B cells typically exhibit high tRNA synthetase expression. Increased ARS expression may be linked to a potential role for the ARS in antigen presentation (154). Not surprisingly, both natural and synthetic tRNA synthetase inhibitors in the context of human cells are immunosuppressive. This property was exploited in the development of aminoacyl-sulfamide IBI derivatives targeting the proliferative skin disease psoriasis (69). Screening efforts to identify compounds that inhibit epidermal growth factor–dependent responses of mouse epidermal cells identified reveromycin, an inhibitor of IleRS, as a hit (155). In a more systematic exploration of this phenomenon, Van de Vijver et al. (156) assessed the immunosuppressive properties of AA-AMS compounds based on all 20 ARS adenylates. These agents inhibited the allogeneic mixed lymphocyte reaction, a clinically approved test used as a proxy to assess transplant rejection. Mupirocin exhibited no immunosuppressive effect up at least 10 μm and served as the negative control. Combinations of multiple AA-AMSs and rapamycin were also tested to look for synergism. When the compounds were arranged on the basis of mixed leukocyte/lymphocyte reaction IC50, the most potent were Asn-AMS, Cys-AMS, and Met-AMS (0.3–0.5 μm), and the least potent were Ile-AMS, Tyr-AMS, and Val-AMS (5.6 μm).
Insights into how ARS inhibitors suppress immune responses have emerged from work demonstrating that natural product inhibitors activate the amino acid starvation pathway. A key observation is that treatment of pro-inflammatory Th17 cells with HF or borrelidin increases the concentration of uncharged (nonaminoacylated) tRNA via inhibition of ARS aminoacylation function. This triggers autophosphorylation of the amino acid starvation sensor kinase GCN2, which phosphorylates the eIF2α initiation factor as part of a generalized activation of the integrated stress response (43, 131, 157, 158). Th17 differentiation is then inhibited, effectively blocking the ability of the host to mount an effective immune response. At higher levels, these compounds can induce apoptosis, which accounts for their toxicity to mammalian cells.
Chronic inflammatory reactions can lead to fibrosis, which is characterized by high levels of production of extracellular matrix proteins (especially collagen) by myofibroblasts. The process is under tight control by TGF-β and is further regulated by matrix metalloproteases (MMPs) and their inhibitors. HF reduces collagen gene expression, and this effect along with stimulation of tissue inhibitor of MMPs explains the ability of HF to reduce fibrosis (159). The biological basis of the HF effect on autoimmune diseases is not fully understood. The specific mechanism may be a consequence of HF's ability to promote the amino acid starvation response. Alternatively, it may involve the phosphorylation of the SMA- and MAD-related protein 3 (SMAD3), thereby inhibiting TGF-β–dependent transcription control. However, no evidence of a direct interaction between HF and SMAD3 has been reported. The discovery of a new class of ProRS inhibitors that is unrelated to HF and yet still exhibits anti-fibrotic activity provides one line of evidence that the anti-fibrotic properties of HF are a direct function of inhibition of ProRS and not the result of interaction with an as yet undefined target (160).
ARS inhibitors: a new frontier for anti-cancer therapies
Noncanonical functions reported for mammalian ARSs include functions as potential cytokines, regulators of angiogenesis, and as regulators of gene expression (161, 162). These novel functions may account for connections between ARSs and cancer that are emerging (163). Although there have been no reports associating specific mutations in ARSs with increased predisposition for cancer, there are initial indications that changes in the expression levels of specific ARSs might increase or decrease the risk of specific cancers. Although a priori one might predict that the levels of all ARSs might rise in fast-growing tumors as a reflection of the need for rapid synthesis of cellular biomass, the emerging literature does not support this view. Instead, specific ARS genes are differentially induced or repressed, depending on the specific cancer.
In one of the first such examinations, elevated levels of TrpRS levels were found to be linked to better survival outcomes in colon cancer (164). To examine possible links between ARSs and cancer more systematically, a bioinformatic survey examined ARSs and ARS-interacting partners for their potential value as prognostic markers of particular cancers (163). The expression profiles and copy number variations in various ARS genes were compared with those of known cancer-associated genes (CAGs) derived from the National Institutes of Health, NCI, cancer gene index. Taking advantage of protein–protein interaction data, the authors then developed a network map linking the various ARSs and ARS-associated proteins to the CAG products, and then arranged the map to emphasize potential Gene Ontology (GO)-associated biological processes. Such maps might serve as a guide to understand emerging regulatory properties of ARSs, which might be strictly dependent on aminoacylation. In a follow-up study applying this approach to glioblastoma multiforme (GBM), expression of several ARSs (ValRS, GlnRS, PheRS, AsnRS, and CysRS) were found to be correlated with improved survival (165). In the second level of analysis, interaction networks featuring these ARSs were defined for several different GBM subtypes; of note, differences in the molecular interactions of CysRS and PheRS were observed for different subtypes. These studies represent an excellent guide for future efforts to mine the large trove of cancer gene expression data and to seek out new links between ARSs and specific cancers.
Alternatively, publicly deposited microarray data can be merged with in-house analyses of ARS expression levels in cancer tissue sections and serum samples to test the role of specific ARSs in cancer. Using this approach, levels of ThrRS (TARS) have been found to be prognostic for risk of ovarian cancer (166) and correlate with a Gleason score (a pathological score for grading tumor aggressiveness) of prostate cancer in the case of prostate cancer. The potential prognostic value of TARS may be linked to its ability to stimulate angiogenesis, described below. More recently, data from mouse model and patient tissues suggest that high MetRS levels may be a marker for increased risk of death from small cell lung cancer (167).
In contrast to the potential value of biomarker studies, there is little evidence to suggest that ARS inhibitors are able to specifically block the proliferation of cancer cells. For example, the anti-ThrRS natural product borrelidin was tested as an inhibitor of the proliferation of malignant ALL cell lines, Jurkat, and CEM cells and showed a greater inhibitory effect on these cell lines relative to fibroblast controls (131, 168). Other cell lines, such as endothelial cells, are quite sensitive to the effects of borrelidin (43, 130). The mechanism of killing likely relies on the integrated stress response described above, because phosphorylation cell cycle arrest and ultimately cell death are heralded by a significant increase in the phosphorylation of the amino acid starvation marker eIF2α (130, 131, 168). If the stress cannot be relieved, apoptosis pathways are triggered. Although the effect of borrelidin on oral cancer cells was initially deemed promising (168), the limited therapeutic window of this compound has discouraged an extensive follow-up on these results.
Another modality by which ARS inhibitors might serve as anticancer therapeutics is by inhibiting one or more of the numerous secondary roles of ARSs that have emerged in recent years (161, 162). Currently, there are relatively few reports of the modulation of these secondary functions by small molecules. One notable example is the interaction of LysRS with the 67-kDa laminin receptor, which participates in cell migration and adhesion (169). Blocking the action of the laminin receptor has the potential to inhibit cancer cell migration, which is essential for metastasis. A small molecule lead compound (BC-K-YH16899) that inhibits the interaction of LysRS and the 67-kDa laminin receptor was identified by HTS and shown to suppress metastasis in different mouse models (170).
Other secondary functions displayed by ARSs that might be critical to cancer development include stimulation of angiogenesis and cell migration (171). Several ARSs appear to modulate angiogenesis, either via stimulation when they are secreted, or as inhibitors. Examples of the former include TyrRS and ThrRS, whereas inhibitory ARS include SerRS and TrpRS (172–176). Earlier work demonstrated that the ThrRS-specific inhibitor borrelidin (previously discussed in the context of malaria) inhibits angiogenesis in both tissue and cellular models (133, 177). The phenotypic effect of BN is the direct result of interactions with ThrRS, which promotes angiogenesis when secreted from human umbilical vein endothelial cells under the influence of TNFα (82). Whereas borrelidin in its native form is toxic to eukaryotic cells, modification of the borrelidin pendant cyclopentane ring can attenuate this toxicity. Thus, BN derivatives like BC-194 possess the ability to modulate angiogenesis without eliciting apoptosis (130, 133, 176). This may ultimately prove useful in the clinic. In an early study involving a mouse model of melanoma, BN was found to reduce the extent of metastasis without influencing tumor volume. In more recent work, an orthotopic mouse model of breast cancer was injected with a liposomal formulation of BN (178). Tumor volume and the number of metastatic nodules were significantly reduced in the BN liposome-treated animals versus controls. These preliminary observations suggest that screening other ThrRS inhibitors that block angiogenesis function for anticancer function might be fruitful.
Summary and future prospects for ARS inhibitors
When bacterial cells are grown in the presence of single ARS inhibitors, resistant mutants arise at a relatively high frequency (∼10−7 m), creating a serious potential liability for using these compounds. In a recent study, Randall et al. (87) sought to estimate the frequency of resistant mutants when cells are treated with two distinct ARS inhibitors (i.e. mupirocin + REP8839). Significantly, the frequency of double mutant isolation was below the limit of detection (<10−12). This indicates that targeting two potentially mutating loci simultaneously is a promising strategy. To achieve this, one effective approach might be to develop a synthetic conjugate that employs a well-characterized uptake moiety (e.g. siderophore or dipeptide) and a toxin “warhead” based on a sulfamide adenosine framework that possesses dual specificity. By exploiting the wealth of current knowledge about ARS amino acid–binding sites, it should be possible to design an amino acid moiety that would simultaneously be compatible with the active sites of multiple hydrophobic ARSs, such as ValRS, IleRS, and LeuRS. One could also devise dual-function inhibitors to simultaneously target AspRS/AsnRS and GluRS/GlnRS. To prevent toxicity, it would be essential to block pathways of uptake of these molecules by human cells.
With regard to efforts to target eukaryotic pathogens, a large collection of promising anti-parasitic ARS lead compounds is now available that target both cytoplasmic and apicoplastid enzymes. Leveraging the extensive library of currently available ARS structures has been a useful tool for in silico docking and rational design efforts, and the hits obtained can be subjected to further SAR to obtain compounds that have improved drug properties and reduced affinity for host cytoplasmic and mitochondrial enzymes. Additionally, rapid sequencing methods have lowered the costs of obtaining complete parasite genomes, allowing targets to be modeled in a relatively rapid fashion. Major challenges remaining include the need to improve selectivity of the parasite enzymes over the host, as well as strategies for combating the relatively rapid evolution of resistance to these inhibitors. As noted above, dual-targeting strategies might be advantageous in limiting the selection of resistant pathogens. In addition, detailed knowledge of structural differences between pathogen and host enzymes may provide a basis to design inhibitors where the development of resistance has an unacceptable fitness cost for the parasite.
In this review, we have focused almost entirely on the potential benefits of reducing disease by inhibiting ARS activity. Significantly, there are also circumstances where compounds that increase ARS activity could have significant clinical value. There is a large and rapidly expanding literature linking mutations in cytoplasmic and mitochondrial ARS genes to neurological diseases of the peripheral and central nervous systems (179). Among the first was a report showing that mutations in the GARS gene encoding GlyRS are linked to Charcot–Marie–Tooth disease (CMT) type 2D and distal spinal muscular atrophy type V (180). Of 38 human ARS genes, mutations associated with human diseases–either largely or partially associated with the nervous system–have been described in a total of 14 cytoplasmic and 17 mitochondrial ARS genes (9). Both autosomal dominant and autosomal recessive forms have been observed, with many of the former conferring CMT2 and many of the latter conferring complex phenotypes featuring brain development defects (e.g. leukoencephalopathy, microcephaly) and multisystem disorders (181). Detailed characterization of the associated mutant proteins indicates that in many (but not all) cases, the mutations confer a loss of function, and the downstream consequences include attenuation of protein synthesis, resulting in abnormal neuron morphology (182, 183). Because of this observed loss of function, it is difficult to envision an inhibitor that would rescue neurological function by decreasing aminoacylation and potential secondary ARS activities. Although there are isolated examples of “gain-of-function” therapeutics for diseases such as cystic fibrosis (184), there are no obvious examples of such compounds for the ARSs. Were compounds to be discovered that could stabilize and perhaps restore at least partial function to mutant ARS enzymes associated with inherited neurological diseases, this could dramatically increase quality of life for a group of patients for whom there are no treatment options. This possibility, along with the many opportunities for therapies based on inhibition of ARS activity, should drive research on these compounds for many years to come.
Acknowledgments
We thank Karen Lounsbury, Alicia Ebert, and Jon Ramsey for advice and suggestions regarding the use ARS inhibitors in the investigation of diverse physiological processes. We also thank Robert Hondal for help in preparation of structures of ARS inhibitors.
This work was supported by National Institutes of Health Grant 5RO1 GM-54899 from NIGMS (to C. S. F.). This is the sixth article in the “tRNAs and aminoacyl-tRNA synthetases in human disease” JBC Reviews series. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ARS
- aminoacyl-tRNA synthetase
- aaS3DA
- N3,C5′-cycloadenosine derivative of the aminoacyl sulfamoyladenosine
- AlaRS
- alanyl-tRNA synthetase
- ArgRS
- arginyl-tRNA synthetase
- AA
- amino acid
- BN
- borrelidin
- CAG
- cancer-associated gene
- CS
- cladosporin
- CysRS
- cysteinyl-tRNA synthetase
- GlyRS
- glycyl-tRNA synthetase
- GluRS
- glutamyl-tRNA synthetase
- Glu-AMS
- glutamyl sulfamoyladenosine
- GlnRS
- glutaminyl-tRNA synthetase
- HF
- halofuginone
- HisRS
- histidyl-tRNA synthetase
- HTS
- high-throughput screening
- IBI
- intermediate-based inhibitor
- IleRS
- isoleucyl-tRNA synthetase
- Ile-AMS
- isoleucyl sulfamoyladenosine
- LeuRS
- leucyl-tRNA synthetase
- LysRS
- lysyl-tRNA synthetase
- MetRS
- methionyl-tRNA synthetase
- PKS
- polyketide synthase
- PMA
- pseudomonic acid
- SAR
- structure–activity relationship
- ThrRS
- threonyl-tRNA synthetase
- TrpRS
- tryptophanyl-tRNA synthetase
- ValRS
- valyl-tRNA synthetase
- FDA
- Food and Drug Administration
- PDB
- Protein Data Bank
- MIC
- minimal inhibitor concentration
- TB
- tuberculosis
- MMP
- matrix metalloprotease
- TGF-β
- transforming growth factor-β
- GBM
- glioblastoma multiforme.
References
- 1. Ling J., Reynolds N., and Ibba M. (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78 10.1146/annurev.micro.091208.073210 [DOI] [PubMed] [Google Scholar]
- 2. Ibba M., and Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 10.1146/annurev.biochem.69.1.617 [DOI] [PubMed] [Google Scholar]
- 3. Ibba M., Francklyn C., and Cusack S., eds (2005) The Aminoacyl-tRNA Synthetases, pp. 1–420, Landes Bioscience, Georgetown, TX [Google Scholar]
- 4. Carter C. W., Jr. (2017) Coding of Class I and II aminoacyl-tRNA synthetases. Adv. Exp. Med. Biol. 966, 103–148 10.1007/5584_2017_93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guo M., and Schimmel P. (2013) Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 9, 145–153 10.1038/nchembio.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin J., Zhou D., Steitz T. A., Polikanov Y. S., and Gagnon M. G. (2018) Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 87, 451–478 10.1146/annurev-biochem-062917-011942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Targoff I. N. (2000) Update on myositis-specific and myositis-associated autoantibodies. Curr. Opin. Rheumatol. 12, 475–481 10.1097/00002281-200011000-00001 [DOI] [PubMed] [Google Scholar]
- 8. Gallay L., Gayed C., and Hervier B. (2018) Antisynthetase syndrome pathogenesis: knowledge and uncertainties. Curr. Opin. Rheumatol. 30, 664–673 10.1097/BOR.0000000000000555 [DOI] [PubMed] [Google Scholar]
- 9. Meyer-Schuman R., and Antonellis A. (2017) Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum. Mol. Genet. 26, R114–R127 10.1093/hmg/ddx231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schimmel P., Tao J., and Hill J. (1998) Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB. J. 12, 1599–1609 10.1096/fasebj.12.15.1599 [DOI] [PubMed] [Google Scholar]
- 11. Finn J., and Tao J. (2003) in The Aminoacyl-tRNA Synthetases (Ibba M., Franckly C., and Cusack S., eds) pp. 405–411, Landes Biosciences, Georgetown, TX [Google Scholar]
- 12. Chenevert R., Bernier S., and Lapointe J. (2003) in Translation Mechanisms (Lapointe J., and Brakier-Gingras L., eds) pp. 416–425, Landes Biosciences Kluwer Academic/Plenum Publishers, Georgetown, TX [Google Scholar]
- 13. Hurdle J. G., O'Neill A. J., and Chopra I. (2005) Prospects for aminoacyl-tRNA synthetase inhibitors as new antimicrobial agents. Antimicrob. Agents Chemother. 49, 4821–4833 10.1128/AAC.49.12.4821-4833.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vondenhoff G. H., and Van Aerschot A. (2011) Aminoacyl-tRNA synthetase inhibitors as potential antibiotics. Eur. J. Med. Chem. 46, 5227–5236 10.1016/j.ejmech.2011.08.049 [DOI] [PubMed] [Google Scholar]
- 15. Dewan V., Reader J. S., and Musier-Forsyth K. (2014) in Aminoacyl-tRNA Synthetases in Biology and Medicine (Kim S., ed) pp. 293–330, Springer, Heidelberg, New York/London [Google Scholar]
- 16. Ibba M., and Söll D. (2001) The renaissance of aminoacyl-tRNA synthesis. EMBO Rep. 2, 382–387 10.1093/embo-reports/kve095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perona J. J., Rould M. A., and Steitz T. A. (1993) Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry 32, 8758–8771 10.1021/bi00085a006 [DOI] [PubMed] [Google Scholar]
- 18. Guth E., Connolly S. H., Bovee M., and Francklyn C. S. (2005) A substrate-assisted concerted mechanism for aminoacylation by a Class II aminoacyl-tRNA synthetase. Biochemistry 44, 3785–3794 10.1021/bi047923h [DOI] [PubMed] [Google Scholar]
- 19. Carter C. W., Jr. (1993) Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 62, 715–748 10.1146/annurev.bi.62.070193.003435 [DOI] [PubMed] [Google Scholar]
- 20. Perona J. J., and Gruic-Sovulj I. (2014) Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top. Curr. Chem. 344, 1–41 [DOI] [PubMed] [Google Scholar]
- 21. Pauling L. (1958) in Festschrift fur Prof. Dr. Arthur Stoll (Birkhauser A., ed) pp. 597–602, Birkhauser Verlag, Basel, Switzerland [Google Scholar]
- 22. Fersht A. R. (1985) Enzyme Structure and Mechanism, pp. 347–368, Freeman, San Francisco [Google Scholar]
- 23. Fersht A. R., and Kaethner M. M. (1976) Enzyme hyperspecificity. Rejection of threonine by the valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry 15, 3342–3346 10.1021/bi00660a026 [DOI] [PubMed] [Google Scholar]
- 24. Fersht A. R. (1977) Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry 16, 1025–1030 10.1021/bi00624a034 [DOI] [PubMed] [Google Scholar]
- 25. Minajigi A., and Francklyn C. S. (2010) Aminoacyl transfer rate dictates choice of editing pathway in threonyl-tRNA synthetase. J. Biol. Chem. 285, 23810–23817 10.1074/jbc.M110.105320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dulic M., Cvetesic N., Perona J. J., and Gruic-Sovulj I. (2010) Partitioning of tRNA-dependent editing between pre- and post-transfer pathways in Class I aminoacyl-tRNA synthetases. J. Biol. Chem. 285, 23799–23809 10.1074/jbc.M110.133553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chong Y. E., Yang X. L., and Schimmel P. (2008) Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J. Biol. Chem. 283, 30073–30078 10.1074/jbc.M805943200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reynolds N. M., Ling J., Roy H., Banerjee R., Repasky S. E., Hamel P., and Ibba M. (2010) Cell-specific differences in the requirements for translation quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 4063–4068 10.1073/pnas.0909640107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hilander T., Zhou X. L., Konovalova S., Zhang F. P., Euro L., Chilov D., Poutanen M., Chihade J., Wang E. D., and Tyynismaa H. (2018) Editing activity for eliminating mischarged tRNAs is essential in mammalian mitochondria. Nucleic Acids Res. 46, 849–860 10.1093/nar/gkx1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J. W., Beebe K., Nangle L. A., Jang J., Longo-Guess C. M., Cook S. A., Davisson M. T., Sundberg J. P., Schimmel P., and Ackerman S. L. (2006) Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration (see comment). Nature 443, 50–55 10.1038/nature05096 [DOI] [PubMed] [Google Scholar]
- 31. Minajigi A., Deng B., and Francklyn C. S. (2011) Fidelity escape by the unnatural amino acid β-hydroxynorvaline: an efficient substrate for Escherichia coli threonyl-tRNA synthetase with toxic effects on growth. Biochemistry 50, 1101–1109 10.1021/bi101360a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song Y., Zhou H., Vo M. N., Shi Y., Nawaz M. H., Vargas-Rodriguez O., Diedrich J. K., Yates J. R., Kishi S., Musier-Forsyth K., and Schimmel P. (2017) Double mimicry evades tRNA synthetase editing by toxic vegetable-sourced non-proteinogenic amino acid. Nat. Commun. 8, 2281 10.1038/s41467-017-02201-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li L., Boniecki M. T., Jaffe J. D., Imai B. S., Yau P. M., Luthey-Schulten Z. A., and Martinis S. A. (2011) Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. U.S.A. 108, 9378–9383 10.1073/pnas.1016460108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Melnikov S. V., Rivera K. D., Ostapenko D., Makarenko A., Sanscrainte N. D., Becnel J. J., Solomon M. J., Texier C., Pappin D. J., and Söll D. (2018) Error-prone protein synthesis in parasites with the smallest eukaryotic genome. Proc. Natl. Acad. Sci. U.S.A. 115, E6245–E6253 10.1073/pnas.1803208115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Curnow A. W., Hong K. W., Yuan R., and Söll D. (1997) tRNA-dependent amino acid transformations. Nucleic Acids Symp. Ser. 1997, 2–4 [PubMed] [Google Scholar]
- 36. Mukai T., Crnković A., Umehara T., Ivanova N. N., Kyrpides N. C., and Söll D. (2017) RNA-dependent cysteine biosynthesis in bacteria and archaea. MBio 8, e00561–17 10.1128/mBio.00561-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Antonellis A., and Green E. D. (2008) The role of aminoacyl-tRNA synthetases in genetic diseases. Annu. Rev. Genomics Hum. Genet. 9, 87–107 10.1146/annurev.genom.9.081307.164204 [DOI] [PubMed] [Google Scholar]
- 38. Bhatt T. K., Kapil C., Khan S., Jairajpuri M. A., Sharma V., Santoni D., Silvestrini F., Pizzi E., and Sharma A. (2009) A genomic glimpse of aminoacyl-tRNA synthetases in malaria parasite Plasmodium falciparum. BMC Genomics 10, 644 10.1186/1471-2164-10-644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ochsner U. A., Sun X., Jarvis T., Critchley I., and Janjic N. (2007) Aminoacyl-tRNA synthetases: essential and still promising targets for new anti-infective agents. Expert Opin. Investig. Drugs 16, 573–593 10.1517/13543784.16.5.573 [DOI] [PubMed] [Google Scholar]
- 40. Osada H., and Isono K. (1985) Mechanism of action and selective toxicity of ascamycin, a nucleoside antibiotic. Antimicrob. Agents Chemother. 27, 230–233 10.1128/AAC.27.2.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scott P. M., Van Walbeek W., and MacLean W. M. (1971) Cladosporin, a new antifungal metabolite from Cladosporium cladosporioides. J. Antibiot. 24, 747–755 10.7164/antibiotics.24.747 [DOI] [PubMed] [Google Scholar]
- 42. Anderton K., and Rickards R. W. (1965) Some structural features of borrelidin, an anti-viral antibiotic. Nature 206, 269 10.1038/206269a0 [DOI] [PubMed] [Google Scholar]
- 43. Keller T. L., Zocco D., Sundrud M. S., Hendrick M., Edenius M., Yum J., Kim Y. J., Lee H. K., Cortese J. F., Wirth D. F., Dignam J. D., Rao A., Yeo C. Y., Mazitschek R., and Whitman M. (2012) Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 8, 311–317 10.1038/nchembio.790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olano C., Wilkinson B., Sánchez C., Moss S. J., Sheridan R., Math V., Weston A. J., Braña A. F., Martin C. J., Oliynyk M., Méndez C., Leadlay P. F., and Salas J. A. (2004) Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tu4055: cluster analysis and assignment of functions. Chem. Biol. 11, 87–97 10.1016/j.chembiol.2003.12.018 [DOI] [PubMed] [Google Scholar]
- 45. Cochrane R. V., Sanichar R., Lambkin G. R., Reiz B., Xu W., Tang Y., and Vederas J. C. (2016) Production of new cladosporin analogues by reconstitution of the polyketide synthases responsible for the biosynthesis of this antimalarial agent. Angew. Chem. Int. Ed. Engl. 55, 664–668 10.1002/anie.201509345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fuller A. T., Mellows G., Woolford M., Banks G. T., Barrow K. D., and Chain E. B. (1971) Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature 234, 416–417 10.1038/234416a0 [DOI] [PubMed] [Google Scholar]
- 47. Agarwal V., and Nair S. K. (2012) Aminoacyl tRNA synthetases as targets for antibiotic development. Med. Chem. Commun. 3, 887–898 10.1039/c2md20032e [DOI] [Google Scholar]
- 48. Gurney R., and Thomas C. M. (2011) Mupirocin: biosynthesis, special features and applications of an antibiotic from a Gram-negative bacterium. Appl. Microbiol. Biotechnol. 90, 11–21 10.1007/s00253-011-3128-3 [DOI] [PubMed] [Google Scholar]
- 49. Silvian L. F., Wang J., and Steitz T. A. (1999) Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupiricin. Science 285, 1074–1077 10.1126/science.285.5430.1074 [DOI] [PubMed] [Google Scholar]
- 50. Nakama T., Nureki O., and Yokoyama S. (2001) Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J. Biol. Chem. 276, 47387–47393 10.1074/jbc.M109089200 [DOI] [PubMed] [Google Scholar]
- 51. Shiozawa H., Kagasaki T., Kinoshita T., Haruyama H., Domon H., Utsui Y., Kodama K., and Takahashi S. (1993) Thiomarinol, a new hybrid antimicrobial antibiotic produced by a marine bacterium. Fermentation, isolation, structure, and antimicrobial activity. J. Antibiot. 46, 1834–1842 10.7164/antibiotics.46.1834 [DOI] [PubMed] [Google Scholar]
- 52. Shiozawa H., Kagasaki T., Torikata A., Tanaka N., Fujimoto K., Hata T., Furukawa Y., and Takahashi S. (1995) Thiomarinols B and C, new antimicrobial antibiotics produced by a marine bacterium. J. Antibiot. 48, 907–909 10.7164/antibiotics.48.907 [DOI] [PubMed] [Google Scholar]
- 53. Gilbart J., Perry C. R., and Slocombe B. (1993) High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob. Agents Chemother. 37, 32–38 10.1128/AAC.37.1.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thomas C. M., Hothersall J., Willis C. L., and Simpson T. J. (2010) Resistance to and synthesis of the antibiotic mupirocin. Nat. Rev. Microbiol. 8, 281–289 10.1038/nrmicro2278 [DOI] [PubMed] [Google Scholar]
- 55. Broom N. J., Cassels R., Cheng H. Y., Elder J. S., Hannan P. C., Masson N., O'Hanlon P. J., Pope A., and Wilson J. M. (1996) The chemistry of pseudomonic acid. 17. Dual-action C-1 oxazole derivatives of pseudomonic acid having an extended spectrum of antibacterial activity. J. Med. Chem. 39, 3596–3600 10.1021/jm950882q [DOI] [PubMed] [Google Scholar]
- 56. Brown M. J., Mensah L. M., Doyle M. L., Broom N. J., Osbourne N., Forrest A. K., Richardson C. M., O'Hanlon P. J., and Pope A. J. (2000) Rational design of femtomolar inhibitors of isoleucyl tRNA synthetase from a binding model for pseudomonic acid-A. Biochemistry 39, 6003–6011 10.1021/bi000148v [DOI] [PubMed] [Google Scholar]
- 57. Class Y. J., and DeShong P. (1995) The pseudomonic acids. Chem. Rev. 95, 1843–1857 10.1021/cr00038a005 [DOI] [Google Scholar]
- 58. Houge-Frydrych C. S., Gilpin M. L., Skett P. W., and Tyler J. W. (2000) SB-203207 and SB-203208, two novel isoleucyl tRNA synthetase inhibitors from a Streptomyces sp. II. Structure determination. J. Antibiot. 53, 364–372 10.7164/antibiotics.53.364 [DOI] [PubMed] [Google Scholar]
- 59. Stefanska A. L., Cassels R., Ready S. J., and Warr S. R. (2000) SB-203207 and SB-203208, two novel isoleucyl tRNA synthetase inhibitors from a Streptomyces sp. I. Fermentation, isolation and properties. J. Antibiot. 53, 357–363 10.7164/antibiotics.53.357 [DOI] [PubMed] [Google Scholar]
- 60. Cassio D., Lemoine F., Waller J. P., Sandrin E., and Boissonnas R. A. (1967) Selective inhibition of aminoacyl ribonucleic acid synthetases by aminoalkyl adenylates. Biochemistry 6, 827–836 10.1021/bi00855a024 [DOI] [PubMed] [Google Scholar]
- 61. Forrest A. K., Jarvest R. L., Mensah L. M., O'Hanlon P. J., Pope A. J., and Sheppard R. J. (2000) Amino adenylate and aminoacyl sulfamate intermediate analogues differing greatly in affinity for their cognate Staphylococcus aureus aminoacyl tRNA synthetases. Bioorg. Med. Chem. Lett. 10, 1871–1874 10.1016/S0960-894X(00)00360-7 [DOI] [PubMed] [Google Scholar]
- 62. Cusack S., Yaremchuk A., and Tukalo M. (1996) The crystal structure of the ternary complex of T. thermophilus seryl-tRNA synthetase with tRNASer and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J. 15, 2834–2842 10.1002/j.1460-2075.1996.tb00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cusack S., Yaremchuk A., and Tukalo M. (1996) The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 15, 6321–6334 10.1002/j.1460-2075.1996.tb01022.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cusack S., Yaremchuk A., and Tukalo M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 19, 2351–2361 10.1093/emboj/19.10.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bovee M. L., Yan W., Sproat B. S., and Francklyn C. S. (1999) tRNA discrimination at the binding step by a Class II aminoacyl-tRNA synthetase. Biochemistry 38, 13725–13735 10.1021/bi991182g [DOI] [PubMed] [Google Scholar]
- 66. Banwell M. G., Crasto C. F., Easton C. J., Forrest A. K., Karoli T., March D. R., Mensah L., Nairn M. R., O'Hanlon P. J., Oldham M. D., and Yue W. (2000) Analogues of SB-203207 as inhibitors of tRNA synthetases. Bioorg. Med. Chem. Lett. 10, 2263–2266 10.1016/S0960-894X(00)00456-X [DOI] [PubMed] [Google Scholar]
- 67. Rath V. L., Silvian L. F., Beijer B., Sproat B. S., and Steitz T. A. (1998) How glutaminyl-tRNA synthetase selects glutamine. Structure 6, 439–449 10.1016/S0969-2126(98)00046-X [DOI] [PubMed] [Google Scholar]
- 68. Bernier S., Dubois D. Y., Habegger-Polomat C., Gagnon L. P., Lapointe J., and Chênevert R. (2005) Glutamylsulfamoyladenosine and pyroglutamylsulfamoyladenosine are competitive inhibitors of E. coli glutamyl-tRNA synthetase. J. Enzyme Inhib. Med. Chem. 20, 61–67 10.1080/14756360400002007 [DOI] [PubMed] [Google Scholar]
- 69. Hill J. M., Yu G., Shue Y. K., Zydowski T. M., and Rebek J. (July 18, 1996) U. S. Patent WO1996US11910
- 70. Ruan B., Nakano H., Tanaka M., Mills J. A., DeVito J. A., Min B., Low K. B., Battista J. R., and Söll D. (2004) Cysteinyl-tRNA(Cys) formation in Methanocaldococcus jannaschii: the mechanism is still unknown. J. Bacteriol. 186, 8–14 10.1128/JB.186.1.8-14.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gadakh B., Smaers S., Rozenski J., Froeyen M., and Van Aerschot A. (2015) 5′-(N-aminoacyl)-sulfonamido-5′-deoxyadenosine: attempts for a stable alternative for aminoacyl-sulfamoyl adenosines as aaRS inhibitors. Eur. J. Med. Chem. 93, 227–236 10.1016/j.ejmech.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 72. Teng M., Hilgers M. T., Cunningham M. L., Borchardt A., Locke J. B., Abraham S., Haley G., Kwan B. P., Hall C., Hough G. W., Shaw K. J., and Finn J. (2013) Identification of bacteria selective threonyl tRNA synthetase (ThrRS) substrate inhibitors by structure-based design. J. Med. Chem. 56, 1748–1760 10.1021/jm301756m [DOI] [PubMed] [Google Scholar]
- 73. Charlton M. H., Aleksis R., Saint-Leger A., Gupta A., Loza E., Ribas de Pouplana L., Kaula I., Gustina D., Madre M., Lola D., Jaudzems K., Edmund G., Randall C. P., Kime L., O'Neill A. J., et al. (2018) N-Leucinyl benzenesulfonamides as structurally simplified leucyl-tRNA synthetase Inhibitors. ACS Med. Chem. Lett. 9, 84–88 10.1021/acsmedchemlett.7b00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang B., De Graef S., Nautiyal M., Pang L., Gadakh B., Froeyen M., Van Mellaert L., Strelkov S. V., Weeks S. D., and Van Aerschot A. (2018) Family-wide analysis of aminoacyl-sulfamoyl-3-deazaadenosine analogues as inhibitors of aminoacyl-tRNA synthetases. Eur. J. Med. Chem. 148, 384–396 10.1016/j.ejmech.2018.02.013 [DOI] [PubMed] [Google Scholar]
- 75. Gadakh B., Vondenhoff G., Lescrinier E., Rozenski J., Froeyen M., and Van Aerschot A. (2014) Base substituted 5′-O-(N-isoleucyl)sulfamoyl nucleoside analogues as potential antibacterial agents. Bioorg. Med. Chem. 22, 2875–2886 10.1016/j.bmc.2014.03.040 [DOI] [PubMed] [Google Scholar]
- 76. Vondenhoff G. H., Gadakh B., Severinov K., and Van Aerschot A. (2012) Microcin C and albomycin analogues with aryl-tetrazole substituents as nucleobase isosters are selective inhibitors of bacterial aminoacyl tRNA synthetases but lack efficient uptake. Chembiochem 13, 1959–1969 10.1002/cbic.201200174 [DOI] [PubMed] [Google Scholar]
- 77. Vondenhoff G. H., Pugach K., Gadakh B., Carlier L., Rozenski J., Froeyen M., Severinov K., and Van Aerschot A. (2013) N-Alkylated aminoacyl sulfamoyladenosines as potential inhibitors of aminoacylation reactions and microcin C analogues containing d-amino acids. PLoS ONE 8, e79234 10.1371/journal.pone.0079234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jarvest R. L., Berge J. M., Houge-Frydrych C. S., Janson C., Mensah L. M., O'Hanlon P. J., Pope A., Saldanha A., and Qiu X. (1999) Interaction of tyrosyl aryl dipeptides with S. aureus tyrosyl tRNA synthetase: inhibition and crystal structure of a complex. Bioorg. Med. Chem. Lett. 9, 2859–2862 10.1016/S0960-894X(99)00488-6 [DOI] [PubMed] [Google Scholar]
- 79. Qiu X., Janson C. A., Smith W. W., Green S. M., McDevitt P., Johanson K., Carter P., Hibbs M., Lewis C., Chalker A., Fosberry A., Lalonde J., Berge J., Brown P., Houge-Frydrych C. S., and Jarvest R. L. (2001) Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors. Protein Sci. 10, 2008–2016 10.1110/ps.18001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pope A. J., McVey M., Fantom K., and Moore K. J. (1998) Effects of substrate and inhibitor binding on proteolysis of isoleucyl-tRNA synthetase from Staphylococcus aureus. J. Biol. Chem. 273, 31702–31706 10.1074/jbc.273.48.31702 [DOI] [PubMed] [Google Scholar]
- 81. Pope A. J., Moore K. J., McVey M., Mensah L., Benson N., Osbourne N., Broom N., Brown M. J., and O'Hanlon P. (1998) Characterization of isoleucyl-tRNA synthetase from Staphylococcus aureus. II. Mechanism of inhibition by reaction intermediate and pseudomonic acid analogues studied using transient and steady-state kinetics. J. Biol. Chem. 273, 31691–31701 10.1074/jbc.273.48.31691 [DOI] [PubMed] [Google Scholar]
- 82. Fang P., Yu X., Jeong S. J., Mirando A., Chen K., Chen X., Kim S., Francklyn C. S., and Guo M. (2015) Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase. Nat. Commun. 6, 6402 10.1038/ncomms7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kim S. Y., Lee Y. S., Kang T., Kim S., and Lee J. (2006) Pharmacophore-based virtual screening: the discovery of novel methionyl-tRNA synthetase inhibitors. Bioorg. Med. Chem. Lett. 16, 4898–4907 10.1016/j.bmcl.2006.06.057 [DOI] [PubMed] [Google Scholar]
- 84. Yu X. Y., Hill J. M., Yu G., Yang Y., Kluge A. F., Keith D., Finn J., Gallant P., Silverman J., and Lim A. (2001) A series of quinoline analogues as potent inhibitors of C. albicans prolyl tRNA synthetase. Bioorg. Med. Chem. Lett. 11, 541–544 10.1016/S0960-894X(00)00697-1 [DOI] [PubMed] [Google Scholar]
- 85. Jarvest R. L., Berge J. M., Berry V., Boyd H. F., Brown M. J., Elder J. S., Forrest A. K., Fosberry A. P., Gentry D. R., Hibbs M. J., Jaworski D. D., O'Hanlon P. J., Pope A. J., Rittenhouse S., Sheppard R. J., et al. (2002) Nanomolar inhibitors of Staphylococcus aureus methionyl tRNA synthetase with potent antibacterial activity against Gram-positive pathogens. J. Med. Chem. 45, 1959–1962 10.1021/jm025502x [DOI] [PubMed] [Google Scholar]
- 86. Critchley I. A., Young C. L., Stone K. C., Ochsner U. A., Guiles J., Tarasow T., and Janjic N. (2005) Antibacterial activity of REP8839, a new antibiotic for topical use. Antimicrob. Agents Chemother. 49, 4247–4252 10.1128/AAC.49.10.4247-4252.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Randall C. P., Rasina D., Jirgensons A., and O'Neill A. J. (2016) Targeting multiple aminoacyl-tRNA synthetases overcomes the resistance liabilities associated with antibacterial inhibitors acting on a single such enzyme. Antimicrob. Agents Chemother. 60, 6359–6361 10.1128/AAC.00674-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kong J., Fang P., Madoux F., Spicer T. P., Scampavia L., Kim S., and Guo M. (2018) High-throughput screening for protein synthesis inhibitors targeting aminoacyl-tRNA synthetases. SLAS Discov. 23, 174–182 10.1177/2472555217734128 [DOI] [PubMed] [Google Scholar]
- 89. Hu Y., Keniry M., Palmer S. O., and Bullard J. M. (2016) Discovery and analysis of natural-product compounds inhibiting protein synthesis in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60, 4820–4829 10.1128/AAC.00800-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hu Y., Palmer S. O., Munoz H., and Bullard J. M. (2014) High throughput screen identifies natural product inhibitor of phenylalanyl-tRNA synthetase from Pseudomonas aeruginosa and Streptococcus pneumoniae. Curr. Drug Discov. Technol. 11, 279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hu Y., Palmer S. O., Robles S. T., Resto T., Dean F. B., and Bullard J. M. (2018) Identification of chemical compounds that inhibit the function of histidyl-tRNA synthetase from Pseudomonas aeruginosa. SLAS Discov. 23, 65–75 10.1177/2472555217722016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Robles S., Hu Y., Resto T., Dean F., and Bullard J. M. (2017) Identification and characterization of a chemical compound that inhibits methionyl-tRNA synthetase from Pseudomonas aeruginosa. Curr. Drug Discov. Technol. 14, 156–168 10.2174/1570163814666170330100238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Guijarro J. I., González-Pastor J. E., Baleux F., San Millán J. L., Castilla M. A., Rico M., Moreno F., and Delepierre M. (1995) Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 270, 23520–23532 10.1074/jbc.270.40.23520 [DOI] [PubMed] [Google Scholar]
- 94. Van de Vijver P., Vondenhoff G. H., Kazakov T. S., Semenova E., Kuznedelov K., Metlitskaya A., Van Aerschot A., and Severinov K. (2009) Synthetic microcin C analogs targeting different aminoacyl-tRNA synthetases. J. Bacteriol. 191, 6273–6280 10.1128/JB.00829-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Metlitskaya A., Kazakov T., Kommer A., Pavlova O., Praetorius-Ibba M., Ibba M., Krasheninnikov I., Kolb V., Khmel I., and Severinov K. (2006) Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J. Biol. Chem. 281, 18033–18042 10.1074/jbc.M513174200 [DOI] [PubMed] [Google Scholar]
- 96. Nocek B., Tikhonov A., Babnigg G., Gu M., Zhou M., Makarova K. S., Vondenhoff G., Van Aerschot A., Kwon K., Anderson W. F., Severinov K., and Joachimiak A. (2012) Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis. J. Mol. Biol. 420, 366–383 10.1016/j.jmb.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kazakov T., Kuznedelov K., Semenova E., Mukhamedyarov D., Datsenko K. A., Metlitskaya A., Vondenhoff G. H., Tikhonov A., Agarwal V., Nair S., Van Aerschot A., and Severinov K. (2014) The RimL transacetylase provides resistance to translation inhibitor microcin C. J. Bacteriol. 196, 3377–3385 10.1128/JB.01584-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Reader J. S., Ordoukhanian P. T., Kim J. G., de Crécy-Lagard V., Hwang I., Farrand S., and Schimmel P. (2005) Major biocontrol of plant tumors targets tRNA synthetase. Science 309, 1533 10.1126/science.1116841 [DOI] [PubMed] [Google Scholar]
- 99. Ataide S. F., and Ibba M. (2006) Small molecules: big players in the evolution of protein synthesis. ACS Chem. Biol. 1, 285–297 10.1021/cb600200k [DOI] [PubMed] [Google Scholar]
- 100. Chopra S., Palencia A., Virus C., Tripathy A., Temple B. R., Velazquez-Campoy A., Cusack S., and Reader J. S. (2013) Plant tumour biocontrol agent employs a tRNA-dependent mechanism to inhibit leucyl-tRNA synthetase. Nat. Commun. 4, 1417 10.1038/ncomms2421 [DOI] [PubMed] [Google Scholar]
- 101. Kim J. G., Park B. K., Kim S. U., Choi D., Nahm B. H., Moon J. S., Reader J. S., Farrand S. K., and Hwang I. (2006) Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc. Natl. Acad. Sci. U.S.A. 103, 8846–8851 10.1073/pnas.0602965103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zeng Y., Kulkarni A., Yang Z., Patil P. B., Zhou W., Chi X., Van Lanen S., and Chen S. (2012) Biosynthesis of albomycin δ(2) provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS Chem. Biol. 7, 1565–1575 10.1021/cb300173x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Benz G., Schroder T., Kurz J., Wunsche C., Karl W., Steffens G., Pfitzner J., and Schmidt D. (1982) Constitution of the deferriform of the albomycins δ1, δ2, and ϵ. Angew. Chem. Int. Ed. Engl. 21, 527–528 10.1002/anie.198205271 [DOI] [Google Scholar]
- 104. Zillmann M., Gorovsky M. A., and Phizicky E. M. (1991) Conserved mechanism of transfer RNA splicing in eukaryotes. Mol. Cell. Biol. 11, 5410–5416 10.1128/MCB.11.11.5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stefanska A. L., Fulston M., Houge-Frydrych C. S., Jones J. J., and Warr S. R. (2000) A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. J. Antibiot. 53, 1346–1353 10.7164/antibiotics.53.1346 [DOI] [PubMed] [Google Scholar]
- 106. Lin Z., Xu X., Zhao S., Yang X., Guo J., Zhang Q., Jing C., Chen S., and He Y. (2018) Total synthesis and antimicrobial evaluation of natural albomycins against clinical pathogens. Nat. Commun. 9, 3445 10.1038/s41467-018-05821-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Baird J. K. (2005) Effectiveness of antimalarial drugs. N. Engl. J. Med. 352, 1565–1577 10.1056/NEJMra043207 [DOI] [PubMed] [Google Scholar]
- 108. Croft S. L., and Olliaro P. (2011) Leishmaniasis chemotherapy–challenges and opportunities. Clin. Microbiol. Infect. 17, 1478–1483 10.1111/j.1469-0691.2011.03630.x [DOI] [PubMed] [Google Scholar]
- 109. Pham J. S., Dawson K. L., Jackson K. E., Lim E. E., Pasaje C. F., Turner K. E., and Ralph S. A. (2014) Aminoacyl-tRNA synthetases as drug targets in eukaryotic parasites. Int. J. Parasitol. Drugs Drug Resist. 4, 1–13 10.1016/j.ijpddr.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rock F. L., Mao W., Yaremchuk A., Tukalo M., Crépin T., Zhou H., Zhang Y. K., Hernandez V., Akama T., Baker S. J., Plattner J. J., Shapiro L., Martinis S. A., Benkovic S. J., Cusack S., and Alley M. R. (2007) An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316, 1759–1761 10.1126/science.1142189 [DOI] [PubMed] [Google Scholar]
- 111. Nare B., Wring S., Bacchi C., Beaudet B., Bowling T., Brun R., Chen D., Ding C., Freund Y., Gaukel E., Hussain A., Jarnagin K., Jenks M., Kaiser M., Mercer L., et al. (2010) Discovery of novel orally bioavailable oxaborole 6-carboxamides that demonstrate cure in a murine model of late-stage central nervous system African trypanosomiasis. Antimicrob. Agents Chemother. 54, 4379–4388 10.1128/AAC.00498-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. O'Dwyer K., Spivak A. T., Ingraham K., Min S., Holmes D. J., Jakielaszek C., Rittenhouse S., Kwan A. L., Livi G. P., Sathe G., Thomas E., Van Horn S., Miller L. A., Twynholm M., Tomayko J., et al. (2015) Bacterial resistance to leucyl-tRNA synthetase inhibitor GSK2251052 develops during treatment of complicated urinary tract infections. Antimicrob. Agents Chemother. 59, 289–298 10.1128/AAC.03774-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Palencia A., Li X., Bu W., Choi W., Ding C. Z., Easom E. E., Feng L., Hernandez V., Houston P., Liu L., Meewan M., Mohan M., Rock F. L., Sexton H., Zhang S., et al. (2016) Discovery of novel oral protein synthesis inhibitors of Mycobacterium tuberculosis that target leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 60, 6271–6280 10.1128/AAC.01339-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Khan S. (2016) Recent advances in the biology and drug targeting of malaria parasite aminoacyl-tRNA synthetases. Malar. J. 15, 203 10.1186/s12936-016-1247-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Saint-Léger A., Sinadinos C., and Ribas de Pouplana L. (2016) The growing pipeline of natural aminoacyl-tRNA synthetase inhibitors for malaria treatment. Bioengineered 7, 60–64 10.1080/21655979.2016.1149270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Milner D. A., Jr. (2018) Malaria pathogenesis. Cold Spring Harb. Perspect. Med. 8 10.1101/cshperspect.a025569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Khan S., Sharma A., Jamwal A., Sharma V., Pole A. K., Thakur K. K., and Sharma A. (2011) Uneven spread of cis- and trans-editing aminoacyl-tRNA synthetase domains within translational compartments of P. falciparum. Sci. Rep. 1, 188 10.1038/srep00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Koepfli J. B., Mead J. F., and Brockman J. A. Jr. (1949) Alkaloids of Dichroa febrifuga; isolation and degradative studies. J. Am. Chem. Soc. 71, 1048–1054 10.1021/ja01171a080 [DOI] [PubMed] [Google Scholar]
- 119. Koepfli J. B., Mead J. F., and Brockman J. A. Jr. (1947) An alkaloid with high antimalarial activity from Dichroa febrifuga. J. Am. Chem. Soc. 69, 1837 10.1021/ja01199a513 [DOI] [PubMed] [Google Scholar]
- 120. Pinion J. L., Bilgili S. F., Eckman M. K., and Hess J. B. (1995) The effects of halofuginone and salinomycin, alone and in combination, on live performance and skin characteristics of broilers. Poult. Sci. 74, 391–397 10.3382/ps.0740391 [DOI] [PubMed] [Google Scholar]
- 121. Zhou H., Sun L., Yang X. L., and Schimmel P. (2013) ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature 494, 121–124 10.1038/nature11774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Geary T. G., Divo A. A., and Jensen J. B. (1989) Stage specific actions of antimalarial drugs on Plasmodium falciparum in culture. Am. J. Trop. Med. Hyg. 40, 240–244 10.4269/ajtmh.1989.40.240 [DOI] [PubMed] [Google Scholar]
- 123. Herman J. D., Pepper L. R., Cortese J. F., Estiu G., Galinsky K., Zuzarte-Luis V., Derbyshire E. R., Ribacke U., Lukens A. K., Santos S. A., Patel V., Clish C. B., Sullivan W. J. Jr., Zhou H., Bopp S. E., et al. (2015) The cytoplasmic prolyl-tRNA synthetase of the malaria parasite is a dual-stage target of febrifugine and its analogs. Sci. Transl. Med. 7, 288ra77 10.1126/scitranslmed.aaa3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Berger J., Jampolsky L. M., and Goldberg M. W. (1949) Borrelidin, a new antibiotic with antiborrelia activity and penicillin enhancement properties. Arch. Biochem. 22, 476–478 [PubMed] [Google Scholar]
- 125. Lumb M., Macey P. E., Spyvee J., Whitmarsh J. M., and Wright R. D. (1965) Isolation of vivomycin and borrelidin, two antibiotics with anti-viral activity, from a species of Streptomyces (C2989). Nature 206, 263–265 10.1038/206263a0 [DOI] [PubMed] [Google Scholar]
- 126. Gao Y. M., Wang X. J., Zhang J., Li M., Liu C. X., An J., Jiang L., and Xiang W. S. (2012) Borrelidin, a potent antifungal agent: insight into the antifungal mechanism against Phytophthora sojae. J. Agric. Food Chem. 60, 9874–9881 10.1021/jf302857x [DOI] [PubMed] [Google Scholar]
- 127. Liu C. X., Zhang J., Wang X. J., Qian P. T., Wang J. D., Gao Y. M., Yan Y. J., Zhang S. Z., Xu P. F., Li W. B., and Xiang W. S. (2012) Antifungal activity of borrelidin produced by a Streptomyces strain isolated from soybean. J. Agric. Food Chem. 60, 1251–1257 10.1021/jf2044982 [DOI] [PubMed] [Google Scholar]
- 128. Otoguro K., Ui H., Ishiyama A., Kobayashi M., Togashi H., Takahashi Y., Masuma R., Tanaka H., Tomoda H., Yamada H., and Omura S. (2003) In vitro and in vivo antimalarial activities of a non-glycosidic 18-membered macrolide antibiotic, borrelidin, against drug-resistant strains of Plasmodia. J. Antibiot. 56, 727–729 10.7164/antibiotics.56.727 [DOI] [PubMed] [Google Scholar]
- 129. Ruan B., Bovee M. L., Sacher M., Stathopoulos C., Poralla K., Francklyn C. S., and Söll D. (2005) A unique hydrophobic cluster near the active site contributes to differences in borrelidin inhibition among threonyl-tRNA synthetases. J. Biol. Chem. 280, 571–577 10.1074/jbc.M411039200 [DOI] [PubMed] [Google Scholar]
- 130. Mirando A. C., Fang P., Williams T. F., Baldor L. C., Howe A. K., Ebert A. M., Wilkinson B., Lounsbury K. M., Guo M., and Francklyn C. S. (2015) Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor. Sci. Rep. 5, 13160 10.1038/srep13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sidhu A., Miller J. R., Tripathi A., Garshott D. M., Brownell A. L., Chiego D. J., Arevang C., Zeng Q., Jackson L. C., Bechler S. A., Callaghan M. U., Yoo G. H., Sethi S., Lin H. S., Callaghan J. H., et al. (2015) Borrelidin Induces the unfolded protein response in oral cancer cells and Chop-dependent apoptosis. ACS Med. Chem. Lett. 6, 1122–1127 10.1021/acsmedchemlett.5b00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ishiyama A., Iwatsuki M., Namatame M., Nishihara-Tsukashima A., Sunazuka T., Takahashi Y., ¯Omura S., and Otoguro K. (2011) Borrelidin, a potent antimalarial: stage-specific inhibition profile of synchronized cultures of Plasmodium falciparum. J. Antibiot. 64, 381–384 10.1038/ja.2011.6 [DOI] [PubMed] [Google Scholar]
- 133. Wilkinson B., Gregory M. A., Moss S. J., Carletti I., Sheridan R. M., Kaja A., Ward M., Olano C., Mendez C., Salas J. A., Leadlay P. F., vanGinckel R., and Zhang M. Q. (2006) Separation of anti-angiogenic and cytotoxic activities of borrelidin by modification at the C17 side chain. Bioorg. Med. Chem. Lett. 16, 5814–5817 10.1016/j.bmcl.2006.08.073 [DOI] [PubMed] [Google Scholar]
- 134. Azcárate I. G., Marín-García P., Camacho N., Pérez-Benavente S., Puyet A., Diez A., Ribas de Pouplana L., and Bautista J. M. (2013) Insights into the preclinical treatment of blood-stage malaria by the antibiotic borrelidin. Br. J. Pharmacol. 169, 645–658 10.1111/bph.12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sugawara A., Tanaka T., Hirose T., Ishiyama A., Iwatsuki M., Takahashi Y., Otoguro K., ¯Omura S., and Sunazuka T. (2013) Borrelidin analogues with antimalarial activity: design, synthesis and biological evaluation against Plasmodium falciparum parasites. Bioorg. Med. Chem. Lett. 23, 2302–2305 10.1016/j.bmcl.2013.02.075 [DOI] [PubMed] [Google Scholar]
- 136. Novoa E. M., Camacho N., Tor A., Wilkinson B., Moss S., Marín-García P., Azcárate I. G., Bautista J. M., Mirando A. C., Francklyn C. S., Varon S., Royo M., Cortés A., and Ribas de Pouplana L. (2014) Analogs of natural aminoacyl-tRNA synthetase inhibitors clear malaria in vivo. Proc. Natl. Acad. Sci. U.S.A. 111, E5508–E5517 10.1073/pnas.1405994111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Schulze C. J., Bray W. M., Loganzo F., Lam M. H., Szal T., Villalobos A., Koehn F. E., and Linington R. G. (2014) Borrelidin B: isolation, biological activity, and implications for nitrile biosynthesis. J. Nat. Prod. 77, 2570–2574 10.1021/np500727g [DOI] [PubMed] [Google Scholar]
- 138. Sun J., Shao J., Sun C., Song Y., Li Q., Lu L., Hu Y., Gui C., Zhang H., and Ju J. (2018) Borrelidins F–I, cytotoxic and cell migration inhibiting agents from mangrove-derived Streptomyces rochei SCSIO ZJ89. Bioorg. Med. Chem. 26, 1488–1494 10.1016/j.bmc.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 139. Curran S. C., Hagen A., Poust S., Chan L. J. G., Garabedian B. M., de Rond T., Baluyot M. J., Vu J. T., Lau A. K., Yuzawa S., Petzold C. J., Katz L., and Keasling J. D. (2018) Probing the flexibility of an iterative modular polyketide synthase with non-native substrates in vitro. ACS Chem. Biol. 13, 2261–2268 10.1021/acschembio.8b00422 [DOI] [PubMed] [Google Scholar]
- 140. Hoepfner D., McNamara C. W., Lim C. S., Studer C., Riedl R., Aust T., McCormack S. L., Plouffe D. M., Meister S., Schuierer S., Plikat U., Hartmann N., Staedtler F., Cotesta S., Schmitt E. K., et al. (2012) Selective and specific inhibition of the Plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe 11, 654–663 10.1016/j.chom.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Khan S., Sharma A., Belrhali H., Yogavel M., and Sharma A. (2014) Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin. J. Struct. Funct. Genomics 15, 63–71 10.1007/s10969-014-9182-1 [DOI] [PubMed] [Google Scholar]
- 142. Istvan E. S., Dharia N. V., Bopp S. E., Gluzman I., Winzeler E. A., and Goldberg D. E. (2011) Validation of isoleucine utilization targets in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 108, 1627–1632 10.1073/pnas.1011560108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Jackson K. E., Pham J. S., Kwek M., De Silva N. S., Allen S. M., Goodman C. D., McFadden G. I., Ribas de Pouplana L., and Ralph S. A. (2012) Dual targeting of aminoacyl-tRNA synthetases to the apicoplast and cytosol in Plasmodium falciparum. Int. J. Parasitol. 42, 177–186 10.1016/j.ijpara.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 144. Kato N., Comer E., Sakata-Kato T., Sharma A., Sharma M., Maetani M., Bastien J., Brancucci N. M., Bittker J. A., Corey V., Clarke D., Derbyshire E. R., Dornan G. L., Duffy S., Eckley S., et al. (2016) Diversity-oriented synthesis yields novel multistage antimalarial inhibitors. Nature 538, 344–349 10.1038/nature19804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kron M., Marquard K., Härtlein M., Price S., and Leberman R. (1995) An immunodominant antigen of Brugia malayi is an asparaginyl-tRNA synthetase. FEBS Lett. 374, 122–124 10.1016/0014-5793(95)01092-S [DOI] [PubMed] [Google Scholar]
- 146. Kron M., Petridis M., Milev Y., Leykam J., and Härtlein M. (2003) Expression, localization and alternative function of cytoplasmic asparaginyl-tRNA synthetase in Brugia malayi. Mol. Biochem. Parasitol 129, 33–39 10.1016/S0166-6851(03)00080-X [DOI] [PubMed] [Google Scholar]
- 147. Sukuru S. C., Crepin T., Milev Y., Marsh L. C., Hill J. B., Anderson R. J., Morris J. C., Rohatgi A., O'Mahony G., Grøtli M., Danel F., Page M. G., Härtlein M., Cusack S., Kron M. A., and Kuhn L. A. (2006) Discovering new classes of Brugia malayi asparaginyl-tRNA synthetase inhibitors and relating specificity to conformational change. J. Comput. Aided Mol. Des. 20, 159–178 10.1007/s10822-006-9043-5 [DOI] [PubMed] [Google Scholar]
- 148. Yu Z., Vodanovic-Jankovic S., Ledeboer N., Huang S. X., Rajski S. R., Kron M., and Shen B. (2011) Tirandamycins from Streptomyces sp. 17944 inhibiting the parasite Brugia malayi asparagine tRNA synthetase. Org. Lett. 13, 2034–2037 10.1021/ol200420u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kalidas S., Cestari I., Monnerat S., Li Q., Regmi S., Hasle N., Labaied M., Parsons M., Stuart K., and Phillips M. A. (2014) Genetic validation of aminoacyl-tRNA synthetases as drug targets in Trypanosoma brucei. Eukaryot. Cell 13, 504–516 10.1128/EC.00017-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Cestari I., and Stuart K. (2013) Inhibition of isoleucyl-tRNA synthetase as a potential treatment for human African trypanosomiasis. J. Biol. Chem. 288, 14256–14263 10.1074/jbc.M112.447441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhao Y., Wang Q., Meng Q., Ding D., Yang H., Gao G., Li D., Zhu W., and Zhou H. (2012) Identification of Trypanosoma brucei leucyl-tRNA synthetase inhibitors by pharmacophore- and docking-based virtual screening and synthesis. Bioorg. Med. Chem. 20, 1240–1250 10.1016/j.bmc.2011.12.035 [DOI] [PubMed] [Google Scholar]
- 152. Zhang N., Zoltner M., Leung K. F., Scullion P., Hutchinson S., Del Pino R. C., Vincent I. M., Zhang Y. K., Freund Y. R., Alley M. R. K., Jacobs R. T., Read K. D., Barrett M. P., Horn D., and Field M. C. (2018) Host-parasite co-metabolic activation of antitrypanosomal aminomethyl-benzoxaboroles. PLoS Pathog. 14, e1006850 10.1371/journal.ppat.1006850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Palencia A., Liu R. J., Lukarska M., Gut J., Bougdour A., Touquet B., Wang E. D., Li X., Alley M. R., Freund Y. R., Rosenthal P. J., Hakimi M. A., and Cusack S. (2016) Cryptosporidium and Toxoplasma parasites are inhibited by a benzoxaborole targeting leucyl-tRNA synthetase. Antimicrob. Agents Chemother. 60, 5817–5827 10.1128/AAC.00873-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Decker W. K., Xing D., Li S., Robinson S. N., Yang H., Steiner D., Komanduri K. V., and Shpall E. J. (2009) Th-1 polarization is regulated by dendritic-cell comparison of MHC Class I and Class II antigens. Blood 113, 4213–4223 10.1182/blood-2008-10-185470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Muguruma H., Yano S., Kakiuchi S., Uehara H., Kawatani M., Osada H., and Sone S. (2005) Reveromycin A inhibits osteolytic bone metastasis of small-cell lung cancer cells, SBC-5, through an antiosteoclastic activity. Clin. Cancer Res. 11, 8822–8828 10.1158/1078-0432.CCR-05-1335 [DOI] [PubMed] [Google Scholar]
- 156. Van de Vijver P., Ostrowski T., Sproat B., Goebels J., Rutgeerts O., Van Aerschot A., Waer M., and Herdewijn P. (2008) Aminoacyl-tRNA synthetase inhibitors as potent and synergistic immunosuppressants. J. Med. Chem. 51, 3020–3029 10.1021/jm8000746 [DOI] [PubMed] [Google Scholar]
- 157. Sundrud M. S., Koralov S. B., Feuerer M., Calado D. P., Kozhaya A. E., Rhule-Smith A., Lefebvre R. E., Unutmaz D., Mazitschek R., Waldner H., Whitman M., Keller T., and Rao A. (2009) Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 324, 1334–1338 10.1126/science.1172638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Kamberov Y. G., Kim J., Mazitschek R., Kuo W. P., and Whitman M. (2011) Microarray profiling reveals the integrated stress response is activated by halofuginone in mammary epithelial cells. BMC Res. Notes 4, 381 10.1186/1756-0500-4-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. McGaha T. L., Phelps R. G., Spiera H., and Bona C. (2002) Halofuginone, an inhibitor of type-I collagen synthesis and skin sclerosis, blocks transforming-growth-factor–β-mediated Smad3 activation in fibroblasts. J. Invest. Dermatol. 118, 461–470 10.1046/j.0022-202x.2001.01690.x [DOI] [PubMed] [Google Scholar]
- 160. Shibata A., Kuno M., Adachi R., Sato Y., Hattori H., Matsuda A., Okuzono Y., Igaki K., Tominari Y., Takagi T., Yabuki M., and Okaniwa M. (2017) Discovery and pharmacological characterization of a new class of prolyl-tRNA synthetase inhibitor for anti-fibrosis therapy. PLoS ONE 12, e0186587 10.1371/journal.pone.0186587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Guo M., Schimmel P., and Yang X. L. (2010) Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 584, 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Guo M., Yang X. L., and Schimmel P. (2010) New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 11, 668–674 10.1038/nrm2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kim S., You S., and Hwang D. (2011) Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat. Rev. Cancer 11, 708–718 10.1038/nrc3124 [DOI] [PubMed] [Google Scholar]
- 164. Ghanipour A., Jirström K., Pontén F., Glimelius B., Påhlman L., and Birgisson H. (2009) The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 18, 2949–2956 10.1158/1055-9965.EPI-09-0456 [DOI] [PubMed] [Google Scholar]
- 165. Kim Y. W., Kwon C., Liu J. L., Kim S. H., and Kim S. (2012) Cancer association study of aminoacyl-tRNA synthetase signaling network in glioblastoma. PLoS ONE 7, e40960 10.1371/journal.pone.0040960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wellman T. L., Eckenstein M., Wong C., Rincon M., Ashikaga T., Mount S. L., Francklyn C. S., and Lounsbury K. M. (2014) Threonyl-tRNA synthetase overexpression correlates with angiogenic markers and progression of human ovarian cancer. BMC Cancer 14, 620 10.1186/1471-2407-14-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Kim E. Y., Jung J. Y., Kim A., Kim K., and Chang Y. S. (2017) Methionyl-tRNA synthetase overexpression is associated with poor clinical outcomes in non-small cell lung cancer. BMC Cancer 17, 467 10.1186/s12885-017-3452-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Habibi D., Ogloff N., Jalili R. B., Yost A., Weng A. P., Ghahary A., and Ong C. J. (2012) Borrelidin, a small molecule nitrile-containing macrolide inhibitor of threonyl-tRNA synthetase, is a potent inducer of apoptosis in acute lymphoblastic leukemia. Invest. New Drugs 30, 1361–1370 10.1007/s10637-011-9700-y [DOI] [PubMed] [Google Scholar]
- 169. Kim D. G., Choi J. W., Lee J. Y., Kim H., Oh Y. S., Lee J. W., Tak Y. K., Song J. M., Razin E., Yun S. H., and Kim S. (2012) Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 26, 4142–4159 10.1096/fj.12-207639 [DOI] [PubMed] [Google Scholar]
- 170. Kim D. G., Lee J. Y., Kwon N. H., Fang P., Zhang Q., Wang J., Young N. L., Guo M., Cho H. Y., Mushtaq A. U., Jeon Y. H., Choi J. W., Han J. M., Kang H. W., Joo J. E., et al. (2014) Chemical inhibition of prometastatic lysyl-tRNA synthetase-laminin receptor interaction. Nat. Chem. Biol. 10, 29–34 10.1038/nchembio.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yao P., and Fox P. L. (2013) Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol. Med. 5, 332–343 10.1002/emmm.201100626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Wakasugi K., Slike B. M., Hood J., Ewalt K. L., Cheresh D. A., and Schimmel P. (2002) Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J. Biol. Chem. 277, 20124–20126 10.1074/jbc.C200126200 [DOI] [PubMed] [Google Scholar]
- 173. Wakasugi K., Slike B. M., Hood J., Otani A., Ewalt K. L., Friedlander M., Cheresh D. A., and Schimmel P. (2002) A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 173–177 10.1073/pnas.012602099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Fukui H., Hanaoka R., and Kawahara A. (2009) Noncanonical activity of seryl-tRNA synthetase is involved in vascular development. Circ. Res. 104, 1253–1259 10.1161/CIRCRESAHA.108.191189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Herzog W., Müller K., Huisken J., and Stainier D. Y. (2009) Genetic evidence for a noncanonical function of seryl-tRNA synthetase in vascular development. Circ. Res. 104, 1260–1266 10.1161/CIRCRESAHA.108.191718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Williams T. F., Mirando A. C., Wilkinson B., Francklyn C. S., and Lounsbury K. M. (2013) Secreted threonyl-tRNA synthetase stimulates endothelial cell migration and angiogenesis. Sci. Rep. 3, 1317 10.1038/srep01317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Wakabayashi T., Kageyama R., Naruse N., Tsukahara N., Funahashi Y., Kitoh K., and Watanabe Y. (1997) Borrelidin is an angiogenesis inhibitor; disruption of angiogenic capillary vessels in a rat aorta matrix culture model. J. Antibiot. 50, 671–676 10.7164/antibiotics.50.671 [DOI] [PubMed] [Google Scholar]
- 178. Funahashi Y., Wakabayashi T., Semba T., Sonoda J., Kitoh K., and Yoshimatsu K. (1999) Establishment of a quantitative mouse dorsal air sac model and its application to evaluate a new angiogenesis inhibitor. Oncol. Res. 11, 319–329 [PubMed] [Google Scholar]
- 179. Tahmasebi S., Khoutorsky A., Mathews M. B., and Sonenberg N. (2018) Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 19, 791–807 10.1038/s41580-018-0034-x [DOI] [PubMed] [Google Scholar]
- 180. Antonellis A., Ellsworth R. E., Sambuughin N., Puls I., Abel A., Lee-Lin S. Q., Jordanova A., Kremensky I., Christodoulou K., Middleton L. T., Sivakumar K., Ionasescu V., Funalot B., Vance J. M., Goldfarb L. G., et al. (2003) Glycyl tRNA synthetase mutations in Charcot–Marie–Tooth disease type 2D and distal spinal muscular atrophy Type V. Am. J. Hum. Genet. 72, 1293–1299 10.1086/375039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Boczonadi V., Jennings M. J., and Horvath R. (2018) The role of tRNA synthetases in neurological and neuromuscular disorders. FEBS Lett. 592, 703–717 10.1002/1873-3468.12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Niehues S., Bussmann J., Steffes G., Erdmann I., Köhrer C., Sun L., Wagner M., Schäfer K., Wang G., Koerdt S. N., Stum M., Jaiswal S., RajBhandary U. L., Thomas U., Aberle H., et al. (2015) Impaired protein translation in Drosophila models for Charcot–Marie–Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun. 6, 7520 10.1038/ncomms8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Tsai P. C., Soong B. W., Mademan I., Huang Y. H., Liu C. R., Hsiao C. T., Wu H. T., Liu T. T., Liu Y. T., Tseng Y. T., Lin K. P., Yang U. C., Chung K. W., Choi B. O., Nicholson G. A., et al. (2017) A recurrent WARS mutation is a novel cause of autosomal dominant distal hereditary motor neuropathy. Brain 140, 1252–1266 10.1093/brain/awx058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. O'Reilly R., and Elphick H. E. (2013) Development, clinical utility, and place of ivacaftor in the treatment of cystic fibrosis. Drug Des. Devel. Ther. 7, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Ubukata M., Osada H., and Isono K. (1985) Synthesis and biological activity of nucleoside antibiotics, ascamycin and its amino acid analogs. Nucleic Acids Symp. Ser. 1985, 81–83 [PubMed] [Google Scholar]
- 186. Zhao C., Qi J., Tao W., He L., Xu W., Chan J., and Deng Z. (2014) Characterization of biosynthetic genes of ascamycin/dealanylascamycin featuring a 5′-O-sulfonamide moiety in Streptomyces sp. JCM9888. PLoS ONE 9, e114722 10.1371/journal.pone.0114722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Werner R. G., Thorpe L. F., Reuter W., and Nierhaus K. H. (1976) Indolmycin inhibits prokaryotic tryptophanyl-tRNA ligase. Eur. J. Biochem. 68, 1–3 10.1111/j.1432-1033.1976.tb10758.x [DOI] [PubMed] [Google Scholar]
- 188. Tanaka K., Tamaki M., and Watanabe S. (1969) Effect of furanomycin on the synthesis of isoleucyl-tRNA. Biochim. Biophys. Acta 195, 244–245 10.1016/0005-2787(69)90621-2 [DOI] [PubMed] [Google Scholar]
- 189. Kohno T., Kohda D., Haruki M., Yokoyama S., and Miyazawa T. (1990) Nonprotein amino acid furanomycin, unlike isoleucine in chemical structure, is charged to isoleucine tRNA by isoleucyl-tRNA synthetase and incorporated into protein. J. Biol. Chem. 265, 6931–6935 [PubMed] [Google Scholar]
- 190. Konrad I., and Röschenthaler R. (1977) Inhibition of phenylalanine tRNA synthetase from Bacillus subtilis by ochratoxin A. FEBS Lett. 83, 341–347 10.1016/0014-5793(77)81037-5 [DOI] [PubMed] [Google Scholar]
- 191. Roth A., Eriani G., Dirheimer G., and Gangloff J. (1993) Kinetic properties of pure overproduced Bacillus subtilis phenylalanyl-tRNA synthetase do not favour its in vivo inhibition by ochratoxin A. FEBS Lett. 326, 87–91 10.1016/0014-5793(93)81767-T [DOI] [PubMed] [Google Scholar]
- 192. Oki T., Hirano M., Tomatsu K., Numata K., and Kamei H. (1989) Cispentacin, a new antifungal antibiotic. II. In vitro and in vivo antifungal activities. J. Antibiot. 42, 1756–1762 10.7164/antibiotics.42.1756 [DOI] [PubMed] [Google Scholar]
- 193. Petraitiene R., Petraitis V., Kelaher A. M., Sarafandi A. A., Mickiene D., Groll A. H., Sein T., Bacher J., and Walsh T. J. (2005) Efficacy, plasma pharmacokinetics, and safety of icofungipen, an inhibitor of Candida isoleucyl-tRNA synthetase, in treatment of experimental disseminated candidiasis in persistently neutropenic rabbits. Antimicrob. Agents Chemother. 49, 2084–2092 10.1128/AAC.49.5.2084-2092.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Heinstein P. (1982) Mechanism of action of granaticin: inhibition of ribosomal RNA maturation and cell cycle specificity. J. Pharm. Sci. 71, 197–200 10.1002/jps.2600710215 [DOI] [PubMed] [Google Scholar]
- 195. Bernier S., Dubois D. Y., Therrien M., Lapointe J., and Chênevert R. (2000) Synthesis of glutaminyl adenylate analogues that are inhibitors of glutaminyl-tRNA synthetase. Bioorg. Med. Chem. Lett. 10, 2441–2444 10.1016/S0960-894X(00)00478-9 [DOI] [PubMed] [Google Scholar]
- 196. Mendes R. E., Alley M. R., Sader H. S., Biedenbach D. J., and Jones R. N. (2013) Potency and spectrum of activity of AN3365, a novel boron-containing protein synthesis inhibitor, tested against clinical isolates of Enterobacteriaceae and nonfermentative Gram-negative bacilli. Antimicrob. Agents Chemother. 57, 2849–2857 10.1128/AAC.00160-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kawamura T., Liu D., Towle M. J., Kageyama R., Tsukahara N., Wakabayashi T., and Littlefield B. A. (2003) Anti-angiogenesis effects of borrelidin are mediated through distinct pathways: threonyl-tRNA synthetase and caspases are independently involved in suppression of proliferation and induction of apoptosis in endothelial cells. J. Antibiot. 56, 709–715 10.7164/antibiotics.56.709 [DOI] [PubMed] [Google Scholar]
- 198. Liu C., Wang X., Yan Y., Wang J., Zhang B., Zhang J., and Xiang W. (2013) Streptomyces heilongjiangensis sp. nov., a novel actinomycete that produces borrelidin isolated from the root surface of soybean [Glycine max (L.) Merr]. Int. J. Syst. Evol. Microbiol. 63, 1030–1036 10.1099/ijs.0.041483-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Nagamitsu T., Takano D., Marumoto K., Fukuda T., Furuya K., Otoguro K., Takeda K., Kuwajima I., Harigaya Y., and Omura S. (2007) Total synthesis of borrelidin. J. Org. Chem. 72, 2744–2756 10.1021/jo062089i [DOI] [PubMed] [Google Scholar]
- 200. Nass G., and Poralla K. (1976) Genetics of borrelidin resistant mutants of Saccharomyces cerevisiae and properties of their threonyl-tRNA-synthetase. Mol. Gen. Genet. 147, 39–43 10.1007/BF00337933 [DOI] [PubMed] [Google Scholar]
- 201. Moss S. J., Carletti I., Olano C., Sheridan R. M., Ward M., Math V., Nur E. A. M., Brana A. F., Zhang M. Q., Leadlay P. F., Mendez C., Salas J. A., and Wilkinson B. (2006) Biosynthesis of the angiogenesis inhibitor borrelidin: directed biosynthesis of novel analogues. Chem. Commun. 2006, 2341–2343 10.1039/b602931k [DOI] [PubMed] [Google Scholar]
- 202. Crasto C. F., Forrest A. K., Karoli T., March D. R., Mensah L., O'Hanlon P. J., Nairn M. R., Oldham M. D., Yue W., Banwell M. G., and Easton C. J. (2003) Synthesis and activity of analogues of the isoleucyl tRNA synthetase inhibitor SB-203207. Bioorg. Med. Chem. 11, 2687–2694 10.1016/S0968-0896(03)00237-2 [DOI] [PubMed] [Google Scholar]