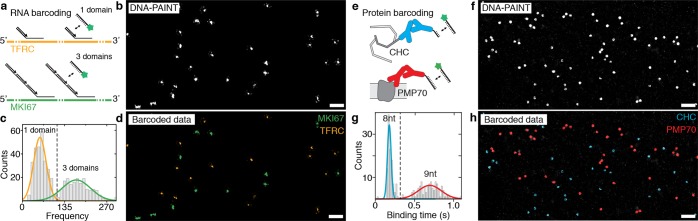
Figure 2.
Engineered binding kinetics allow simultaneous multiplexed super-resolution imaging of RNA and proteins in cells. (a) Scheme showing the implementation of frequency barcoding for smRNA-FISH. Two distinct RNA species (TFRC and MKI67) are labeled with FISH probes featuring 40 binding sites for DNA-PAINT or 120 binding sites, respectively. (b) Resulting DNA-PAINT data after image acquisition shows TFRC and MKI67 mRNA molecules as single spots, which are not yet distinguishable. (c) Plotting the blinking frequency for all detected single mRNA molecules shows a clearly distinguishable distribution of a low and a high frequency, corresponding to the FISH probe set for TFRC (yellow) and MKI67 (green), respectively. (d) Distinct frequencies are used to assign a pseudocolor for each RNA species. (e) Scheme showing the implementation of duration barcoding for protein detection. Two distinct protein species are labeled with DNA-conjugated antibodies featuring an 8 and 9 nt binding site for DNA-PAINT imaging. (f) Resulting DNA-PAINT data after image acquisition shows CHC and PMP70 proteins as clusters, which are not yet distinguishable. (g) Plotting the binding duration for selected protein locations shows a clearly distinguishable distribution of short and long binding species, corresponding to the two proteins. (h) Distinct durations are used to assign a pseudocolor for each protein species. Scale bars: 1 μm.