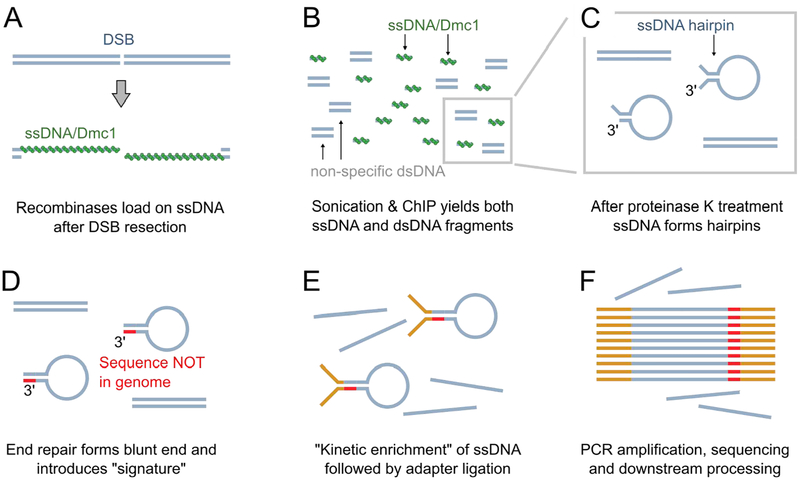
Figure 1. ChIP-SSDS schematic.
(A) ChIP-SSDS captures ssDNA bound by meiotic recombinases to map DSB hotspot locations. (B) After fixation and sonication, DMC1-bound ssDNA is immunoprecipitated using an anti-DMC1 antibody. (C) Deproteinized ssDNA will spontaneously form hairpins, mediated by intra-molecular micro-homologies. (D) The 3’ ends of hairpins are trimmed, then extended, using the 5’ end of the ssDNA molecule as a template. This both introduces a “signature” that allows downstream identification of ssDNA-derived sequencing reads, and ensures that hairpins have double stranded ends. (E) The DNA is denatured, then allowed to return to room temperature. In the reaction timeframe, ssDNA hairpins will re-form because of fast intra-molecular reannealing, however dsDNA will not renature. Sequencing adapters are then ligated to blunt-ended DNA, which will primarily occur at ssDNA-derived hairpins. (F) DNA with ligated adapters is then amplified and sequenced. A computational pipeline then specifically uses the “signature” sequence (D) to identify ssDNA.