SUMMARY
Double homeobox (DUX) transcription factors are unique to eutherian mammals. DUX4 regulates expression of repetitive elements during early embryogenesis, but misexpression of DUX4 causes facioscapulohumeral muscular dystrophy (FSHD) and translocations overexpressing the DUX4 double homeodomain cause B cell leukemia. Here, we report the crystal structure of the tandem homeodo-mains of DUX4 bound to DNA. The homeodomains bind DNA in a head-to-head fashion, with the linker making anchoring DNA minor-groove interactions and unique protein contacts. Remarkably, despite being tandem duplicates, the DUX4 homeodomains recognize different core sequences. This results from an arginine-to-glutamate mutation, unique to primates, causing alternative positioning of a key arginine side chain in the recognition helix. Mutational studies demonstrate that this primate-specific change is responsible for the divergence in sequence recognition that likely drove coevolution of embryonically regulated repeats in primates. Our work provides a framework for understanding the endogenous function of DUX4 and its role in FSHD and cancer.
In Brief
Lee et al. determine the crystal structure of the facioscapulohumeral muscular dystrophy and cancer-associated transcription factor DUX4, bound to DNA. The structure gives insight into how the double homeodomain of DUX4, which is related by duplication of an ancestral homeodomain, has evolved different sequence specificities, uniquely in the primate lineage.
Graphical Abstract
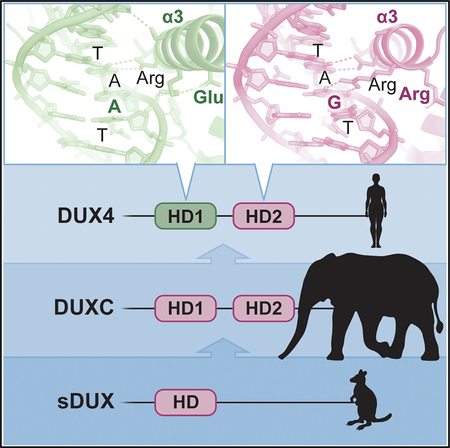
INTRODUCTION
Transcription factors are highly modular proteins, not uncommonly containing more than one DNA-binding domain; PAX family transcription factors, for example, contain both a homeo-domain and a Paired domain. The homeodomain is an ancient motif, being present in eukaryotes as diverse as yeast and vertebrates. Although homeodomains typically bind DNA as dimers, a tandem homeodomain architecture within a single protein is highly unusual: there are no double homeodomain proteins in species outside of placental mammals, and within mammals, the function of the DUX (double homeobox) family is enigmatic (Clapp et al., 2007). The DUX family seems to have radiated out following a mutation in the progenitor to eutherian mammals, in which part of a gene containing a single homeobox (Leidenroth and Hewitt, 2010) was duplicated such that the gene now contained two homeoboxes in tandem, connected by a short linker. Little is known about the mode of DNA interaction of such double homeodomain proteins, in particular whether they bind DNA in tandem (head-to-tail) or symmetrically about a dyad axis (head to head).
One of the descendants of the original DUX gene is DUX4, which in humans is present in a multicopy array near the telo-mere of chromosome 4 (Gabriëls et al., 1999). DUX4 seems to be normally expressed in testis (Snider et al., 2010) and in cleavage-stage embryos (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017); however, misexpression via gain-of-function mutations is implicated in two distinct human diseases: the genetic myopathy, facioscapulohumeral muscular dystrophy (FSHD) (Gabriëls et al., 1999; Lemmers et al., 2010; Wijmenga et al., 1992), and B cell leukemia (Yasuda et al., 2016; Zhang et al., 2016a). In the case of FSHD, overexpression of the full-length protein is implicated: its expression is observed at low levels in cultured myoblasts derived from patient biopsies (Block et al., 2013; Snider et al., 2010). Low-level full-length DUX4 protein expression interferes with myogenesis in vitro and in vivo (Bosnakovski et al., 2017; Bosnakovski et al., 2008b; Dandapat et al., 2014), while high-level DUX4 protein expression is toxic to myoblasts (Bosnakovski et al., 2008b), including endogenous DUX4 in human FSHD myoblasts (Rickard et al., 2015), as well as other cell types (Kowaljow et al., 2007). This toxicity is unique to DUX4 within the human DUX family. Toxicity of DUX4 is conferred by its distinct C terminus (Bosnakovski et al., 2008a), which interacts with p300 to strongly activate expression of its target genes (Choi et al., 2016). Interestingly, the translocations that lead to B cell leukemia always lead to B cell-specific overexpression of a mutant version of DUX4, which contains the double homeodomain motif but lacks the toxicity-associated C-terminal transactivation domain; indeed, the full-length DUX4 protein is actually toxic to B cells (Yasuda et al., 2016). Thus, although these two diseases must be caused by different fundamental transcriptional mechanisms, both absolutely require the specific DNA-binding properties of DUX4. Understanding the principles of DNA recognition by the DUX4 double homeodomain thus provides mechanistic insight into both the natural function of DUX4 in eutherian mammals, as well as the pathological function in FSHD and B cell leukemia.
The two homeodomains of DUX4 belong to the Paired-homeobox (PAX) branch of homeodomain family, but they are more similar to each other in amino acid sequence than to the homeo-domains of any other PAX family members, as expected of an internal duplication within the ancestral PAX-related gene (Leiden-roth and Hewitt, 2010). PAX homeodomains typically bind as head-to-head dimers over a TAAT core (Birrane et al., 2009). The gap between the TAAT and ATTA may be 2 nucleotides (i.e., 5′-TAATNNATTA-3′, a palindromic “P2” site), as in the case of PAX7 (Soleimani et al., 2012), or 3 nucleotides (i.e., 5′-TAATNNNATTA-3′, a P3 site), as in the case of Drosophila Paired (Wilson et al., 1995). However, the DUX4 consensus motif is quite distinct: 5′-TAATCTAATCA-3′ (Geng et al., 2012; Zhang et al., 2016b). Assuming the highly related homeodomains recognize the same core, one could envision the DUX4 homeo-domains recognizing TAAT in tandem on the same strand (5′-TAATCTAATCA-3′) forcing them into a head-to-tail dimer configuration. This would be similar to the modes of DNA binding by Even-skipped homeodomain (Hirsch and Aggarwal, 1995) and the Hox-Pbx hetero-dimeric homeodomains (LaRonde-LeBlanc and Wolberger, 2003; Passner et al., 1999; Piper et al., 1999) (Figure S1A) and was suggested in a recent structural study of the isolated second homeodomain of DUX4 (Dong et al., 2018). However, this latter structure does not actually show a home-odomain positioned over the TAAT core, and therefore its significance remains unclear (Aihara et al., 2018). Alternatively, if core sequence preference has diverged such that one homeodomain recognizes TGAT, the DUX4 consensus could be viewed as a non-palindrome (N3: 5′-TAATCTAATCA-3′) in which the inverted core sequences separated by 3 bp are recognized by the tandem homeodomains in a head-to-head orientation.
RESULTS AND DISCUSSION
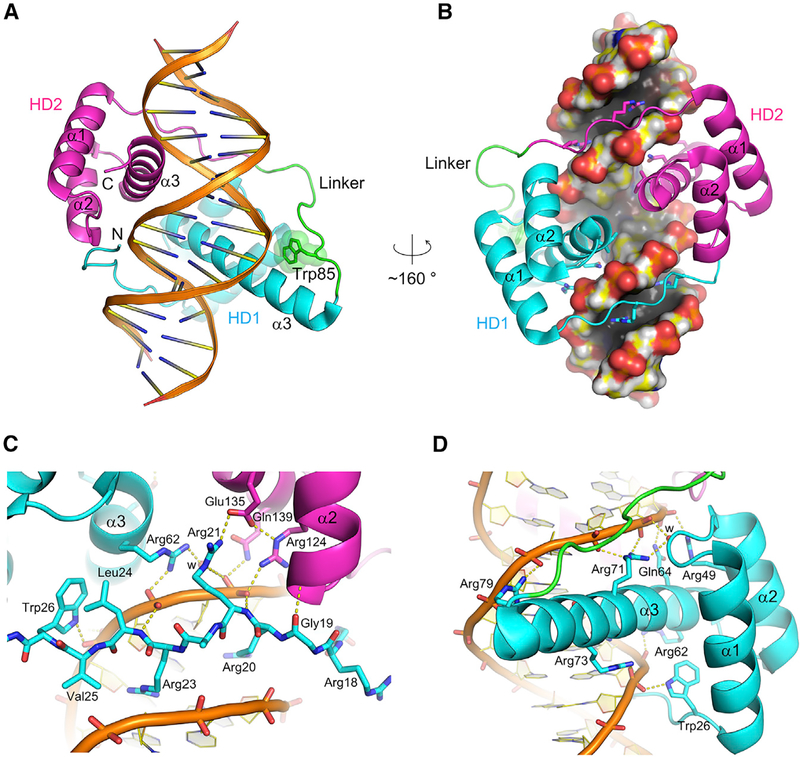
To address how the DUX4 double homeodomain binds DNA, we have crystallized and solved the structure of the DUX4 N terminus (residues 15–155, which includes the double homeodo-main), in complex with a DNA substrate including the consensus motif. The structure refined to 2.12-Å resolution shows that each homeodomain (HD1 and HD2) of DUX4 has the canonical three α-helical bundle architecture, connected through a well-ordered linker segment that plays critical roles in positioning the two domains by making both protein and DNA contacts (Figure 1). The two homeodomains bound to DNA are arranged in a head-to-head fashion and are related by a dyad of the pseudo-palindromic target sequence, although α3 of HD1 is significantly longer than that of HD2. Thus, DUX4 recognizes its target sequence as an inverted repeat, where HD1 and HD2 recognize different sequences, 5′-TAAT-3′and 5′-TGAT-3′, respectiveely. As observed in other structurally characterized homeodomain-DNA complexes (Hirsch and Aggarwal, 1995; Kissinger et al., 1990; Li et al., 1995; Passner et al., 1999; Piper et al., 1999; Wilson et al., 1995) and for DUX4 HD2 (Aihara et al., 2018; Dong et al., 2018), the sequence readout by each homeodomain of DUX4 involves interactions in both the major and minor grooves of DNA. The third α-helix (α3) of both HD1 and HD2 is inserted into the DNA major groove for direct base contacts (Figures 1B, 1D, 2A, and 2B). On either side of the major groove harboring the α3 helices, the arginine-rich stretches N-terminal to the first helix (α1) of HD1 and HD2 traverse the minor grooves (Figures 1B and 1C). These interactions allow the DUX4 polypeptide comprising the tandem homeodomains to follow a circular path to span 3 consecutive grooves on one face of DNA, effectively clamping down the target DNA molecule (Figures 1A and 1B). This topology is distinct from those observed previously for other transcription factors containing linked α-helical DNA-binding domains, such as Oct-1 POU domain (Klemm et al., 1994) or yeast Reb1 (Jaiswal et al., 2016). In an electrophoretic mobility shift assay (EMSA), formation of a stable DUX4(15–155)-DNA complex required the binding sites for both homeodo-mains that are separated ideally by 3 bp (Figures S2A and S2B), consistent with the crystal structure.
Figure 1. Overview of the DUX4(15–155)-DNA Complex.
(A and B) Overall structure of the DUX4 double homeodomain bound to DNA. DUX4 HD1 and HD2 are colored in cyan and magenta, respectively, with the intervening region including Trp85 in green. (A) The DNA backbones and base-pairs are depicted by ribbons and ladders, respectively.
(B) The molecular surface for DNA is shown. Side chains are shown in sticks for the arginines that insert deep into the minor groove and the residues from α3 that make sequence-specific contacts in the major groove.
(C) DNA minor-groove contacts made by the N-terminal arginine-rich stretch preceding α1 of HD1.
(D) Sequence-independent contacts in the DNA major groove by HD1. Although HD1 and HD2 of DUX4 make similar DNA backbone contacts, HD1 shows some unique interactions, including the backbone contact made by Trp26 at the end of the N-terminal stretch and that by Arg79 at the end of the longer α3 helix. Hydrogen-bonding interactions are depicted by yellow dotted lines. Water molecules mediating protein-DNA interactions are represented by small red spheres and labeled “w.”
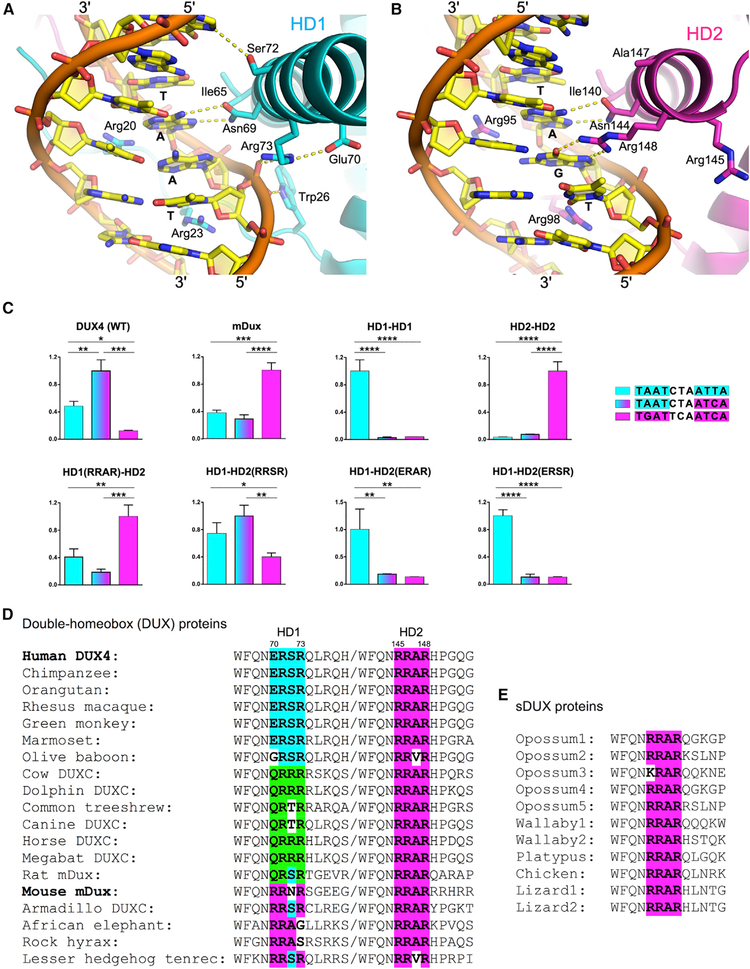
Figure 2. Sequence-Specificity Determinants.
(A and B) Views of the α3 helices of HD1 (A) and HD2 (B) inserted in the DNA major groove, with the side chains making base-specific contacts and some of the surrounding residues shown in sticks. Hydrogen-bonding interactions are depicted by yellow dotted lines. Note differential positioning of HD1 Arg73 (ERSR) and HD2 Arg148 (RRAR).
(C) Target DNA sequence preference of DUX4 HD mutants. Luciferase assays measuring transcriptional activation by various mutants of DUX4. Each protein construct was tested in parallel against reporters bearing two copies of the three target DNA sequences (indicated). Expression levels normalized against the highest of the three are plotted. The mean + SEM of 3 independent experiments is plotted. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by one-way ANOVA analysis with Tukey’s multiple-comparisons test.
(D and E) Sequence alignment of the residues from double-homeobox (DUX) (D) and single-homeobox (sDUX) (E) family proteins, corresponding to DUX4 Trp66 ~His78 in HD1 and Trp141~ Gly153 in HD2. The region that determines the target DNA sequence preference of each homeodomain is shown in bold. The DUX4 HD1-type sequence is highlighted in cyan, whereas the DUX4 HD2 (sDUX)-type sequence is in magenta and the third class (QRxR) is in green. This coloring scheme is also used for the histograms in (C). Protein sequences are from Uniprot or Leidenroth and Hewitt (2010).
The overall structure of the DUX4(15–155)-DNA complex is similar to that of the Drosophila Paired (S50Q) homodimer bound to a P3 DNA site (Wilson et al., 1995), including the ~20°bending and narrowing of the minor grooves of DNA toward the protein (Figure S1B). The sequence recognition in the major groove by α3 of HD1 involves hydrogen bonds and van der Waals contacts by Asn69 and Ile65, respectively, with two bases (in bold and italics) of the 5′-TAAT-3′core (Figure 2A). Asn144 and Ile140 of HD2 make similar contacts with two bases of 5′-TGAT-3′ (Figure 2B). These interactions are conserved among PAX family homeodomains (Asn51 and Val47 of PAX3 [Birrane et al., 2009]; Figure S3), although Ile65 and Ile140 of DUX4, in place of the highly conserved Val in PAX proteins, make closer van der Waals contact with the thymine 5-methyl group. Arg20 and Arg23 from the N terminus of HD1 insert deep into the adjacent minor groove to hydrogen bond with thymine O2 and adenine N3 atoms of 5′-TAAT-3′ (Figure 2A). An intervening residue Arg21 forms a salt bridge with Glu135 from HD2 over the DNA strand, while the surrounding main chain amide groups make phosphate backbone contacts with either DNA strand to help position the N-terminal segment (Figure 1C). Arg95 and Arg98 of HD2 similarly interact with 5′-TGAT-3′ in the minor groove (Figure 2B), and the intervening Arg96 reciprocally interacts with Glu60 from HD1. The insertion of arginine side chains into a compressed DNA minor groove is a hallmark feature of AT-rich sequence recognition by homeo-domains (Slattery et al., 2014). While many of the DNA backbone contacts are conserved between HD1 and HD2 as well as between DUX4 and other PAX homeodomains, DUX4 shows unique modes of interaction. These include a DNA phosphate contact made by Trp26 (corresponding to Phe in most PAX family members and Val in DUX4 HD2; Figures S3 and S4) preceding α1 of HD1, and that made by Arg79 near the C-terminal end of the long α3 of HD1 with the opposing DNA strand (Figures 1C, 1D, and 2A).
The remarkable implication of this structure is that, despite sharing high structural similarity and being more related in amino acid sequence to each other than to any other homeodo-mains, HD1 and HD2 of DUX4 exhibit different target DNA sequence preferences. Our structure shows that the key determinant is Arg148 of HD2, which forms bidentate hydrogen bonds with the guanine base of 5′-TGAT-3′ in the major groove (Figures 2B and S4). A similar arginine-guanine interaction was observed for other homeodomains with 5′-TGAT-3′ target preference, including yeast MATa1(PDB: 1YRN) (Li et al., 1995), Drosophila Extradenticle (PDB: 1B8I) (Passner et al., 1999), and human PBX1 (PDB: 1PUF) (LaRonde-LeBlanc and Wolberger, 2003). Curiously, arginine at this position is conserved in DUX4 HD1 (Arg73) as well as in PAX homeodo-mains (Arg55 in PAX3 [Birrane et al., 2009]; Figure S3), both of which recognize the 5′-TAAT-3′ core sequence. However, Arg73 of HD1 does not project into the major groove and instead makes a backbone phosphate contact (Figure 2A). Likewise, Arg55 of PAX3 is pointed away from DNA (Birrane et al., 2009). Thus, it is not the mere presence of arginine but its positioning that confers unique sequence preference.
To understand this unexpected structural divergence, we compared the DNA interfaces of HD1 and HD2. Two factors differentiate HD1 and HD2 that may contribute to the differential positioning of this critical arginine. First, a neighboring residue Glu70 forms a salt bridge with Arg73 in HD1, keeping the argi-nine side chain at bay (Figure 2A). PAX3 Arg55 is similarly bonded with Glu17 from α1 (Birrane et al., 2009). In contrast, the residue corresponding to Glu70 in HD2 is Arg145, which does not attract Arg148 (Figure 2B). The second factor is the longer α3 helix of HD1. Although the positionings of α3 of HD1 and HD2 relative to DNA are very similar, the Cα-Cβ bond vector of Arg73 does not point toward the DNA major groove, which precludes direct base contacts by Arg73 even with side chain torsion (χ) angles adjustment. In contrast, α3 is interrupted immediately following Arg148 in HD2, and accordingly the main chain carbonyl group of Arg148 is not hydrogen bonded. This provides more flexibility for the HD2 arginine residue so its side chain can point straight toward the guanine base of the 5′-TGAT-3′ motif (Figure 2B).
To interrogate these two potential explanations for distinct sequence selectivity of HD1 and HD2, we mutated specific residues of HD1 and HD2 and tested the ability of the mutant DUX4 proteins to activate luciferase reporters with all 3 possible configurations of TAAT and TGAT cores (i.e., palindromic P3 TAAT, palindromic P3 TGAT, or pseudo-palindromic N3 TAAT/TGAT) (Figure 2C). The luciferase reporter assay using full-length DUX4 constructs is a more stringent functional test of the effects of protein or DNA sequence alterations than EMSA with the DUX4 double homeodomain (Figure S2). Based on the sequences preceding the key arginine residues Arg73 in HD1 (68QNERSR73) and Arg148 in HD2 (143QNRRAR148) (Figures 2D and S3), we made various changes in the α3 of HD2 to mimic HD1: HD2-ERAR (R145E), HD2-RRSR (A147S), and HD2-ERSR (R145E/A147S). We also made a key reciprocal change in HD1 to mimic HD2 (HD1-RRAR: E70R/S72A), and complete homeodo-main replacements (HD1HD1 or HD2HD2) for reference. Ser72 of HD1, which corresponds to Ala147 of HD2, forms a weak hydrogen bond (~3.5Å between non-hydrogen atoms) with the guanine N7 atom from the CG pair within the 3-bp spacer region (5′-TAATCTAATCA-3′) (Figure 2A). In addition, we substituted residues near the end of α3 in HD1 (74QLRQHR79) for the corresponding residues in HD2 (149HPGQGG154) to generate a “long helix” HD1-like version of HD2.
As expected, the wild-type DUX4 showed strong preference for the 5′-TAATCTAATCA-3′ target and a lower activity on 5′-TAATCTAATTA-3′. The latter becomes the preferred target after a complete replacement of HD2 with HD1 (Figure 2C). The R145E substitution conferred a similar effect to swapping the entire HD, albeit somewhat less potently, and A147S alone allowed DUX4 to recognize both targets comparably. Further combining R145E and A147S led to a complete reversal of the preference, rendering DUX4 to selectively bind to a canonical P3-type PAX target sequence 5′-TAATCTAATTA-3′ as “HD1HD1.” Changes downstream of Arg148 to mimic the longer α3 of HD1 (long helix) did not have a significant effect (data not shown). These results confirm that Arg145 in HD2, in place of Glu70 in HD1, is important for the unique preference of HD2 for 5′-TGAT-3′. Mouse Dux (mDux), which has RRNR and RRAR at this position in HD1 and HD2, respectively (Figure 2D), was reported to have the canonical target sequence of 5′-TGATTCAATCA-3′ (Eidahl et al., 2016; Whiddon et al., 2017). Consistent with the above results, HD1-RRAR, HD2HD2, and mDux constructs all showed strong preference for the canonical mDux target sequence (Figure 2C). Interestingly, we found that Ala147 in HD2 as opposed to Ser72 in HD1 contributes to the preference of HD2 for 5′-TGAT-3′, suggesting that the conformation of the neighboring protein or DNA residues also has a significant effect on the sequence recognition. While DUX4(15–155) R145E/A147S did not show a dramatic defect in the protein-DNA complex formation in EMSA, it bound particularly poorly to the N2 site, likely due to a combination of compromised DNA-binding by HD2 and a wrong (2 bp) spacing between the two core sequences (Figure S2C).
The critical role of the 145RRAR148 stretch including Arg145 in determining the target sequence preference of DUX4 suggests that the “single-homeodomain DUX” (sDUX) protein found in non-placental mammals, which likely represents the progenitor of DUX4 and has RRAR as in HD2 of DUX4 (Leidenroth and Hewitt, 2010) (Figure 2E), would also preferentially bind 5′-TGAT-3′. Accordingly, the ancestral DUX protein generated via gene duplication would have preferentially recognized a 5′-TGAT—ATCA-3′ target sequence, as does mDUX (Figure 2C). Notably, a comparison between the amino acid sequences of mammalian DUX4 orthologs show strict conservation of the RRAR stretch of HD2, but considerable variation in the corresponding position in HD1. Whereas most primate DUX4 sequences share “ERSR” with human DUX4 (70ERSR73), DUX4/DUXC from several other mammals have QRxR, and mDux has RRNR (Figure 2D). This predicts that, outside of primates, HD1 and HD2 of DUXC would prefer the same core sequence: TGAT. Thus, a unique sequence specificity for DUX4 distinguishes primates from the rest of mammals.
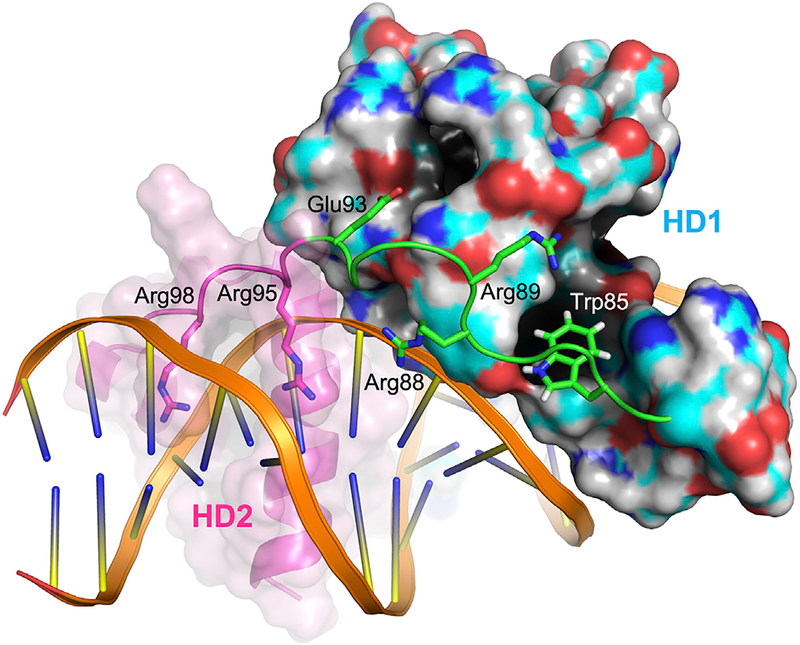
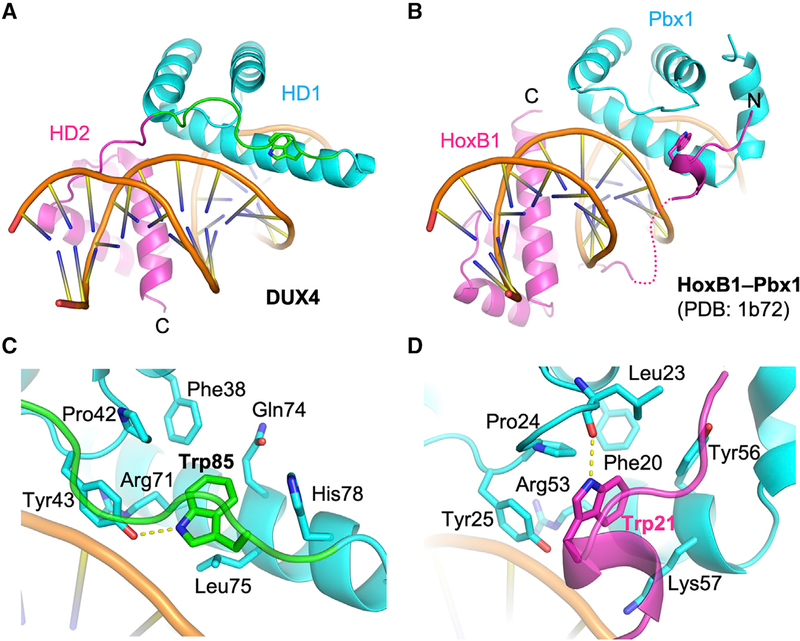
A superposition of the DUX4(15–155)-DNA and Drosophila Paired homodimer-DNA complexes shows a notable deviation of the two structures near the end of the recognition helix (α3) of DUX4 HD1, which harbors Arg79 mentioned above (Figure S1B). The α3 helix of HD1 is curved toward the linker (82SRPWPGRRGPPEG94) connecting HD1 and HD2 of DUX4. The presence of a linker, which constrains positioning of HD1 and HD2, is a unique structural feature of DUX proteins containing tandemly linked homeodomains. The indole ring of Trp85 from this stretch (in bold and italic) docks into a hydrophobic pocket lined by Phe38, Pro42, Tyr43, Gln74, Lue75, and His78 from HD1, where its NεH group is hydrogen bonded to Tyr43 (Figures 1A, 3, 4A, and 4C). The proline-rich linker segment anchored by Trp85 makes extensive contacts with HD1, including a hydrogen bond between Glu93 and Thr48. In addition, Arg88 from the linker also makes a DNA backbone contact. Through these interactions, the linker likely facilitates cooperative DNA binding by HD1 and HD2. A notable analog to this mode of interaction exists in the Hox-Pbx1 (LaRonde-LeBlanc and Wolberger, 2003; Piper et al., 1999) (Ubx-Exd in Drosophila [Passner et al., 1999]) heterodimer-DNA complexes: the “YPWM” motif in the N terminus of the Hox homeodomain mediates similar interactions with Pbx1, where the tryptophan residue (in bold and italic) docks into a pocket formed near the C terminus of Pbx1 (Figures 4B and 4D). However, the mode of homeodo-main dimerization in the head-to-tail Hox-Pbx1 heterodimers is fundamentally different from the head-to-head configuration for DUX4 HD1-HD2, and the interactions of the peptide motifs including the tryptophan residues are locally not conserved between the Hox N terminus and the DUX4 linker (Figures 4C and 4D). Thus, involvement of similar binding pockets at a common location may have resulted from convergent evolution.
Figure 3. Linker Interactions.
The inter-domain linker connecting HD1 and HD2 of DUX4 is shown in green. Trp85 docks into a hydrophobic pocket on the HD1 surface. Other residues from the linker also make protein or DNA contacts to coordinate positioning of the two homeodomains. Solid and transparent surfaces are shown for HD1 and HD2, respectively.
Figure 4. Comparison between DUX4 and HoxB1-Pbx1 Heterodimer.
(A and B) A side-by-side comparison between DUX4 (A) and HoxB1-Pbx1 heterodimer (B) (LaRonde-LeBlanc and Wolberger, 2003; Piper et al., 1999) bound to respective DNA substrate, highlighting similarity in the positioning of a key Trp residue in coordinating the two homeodomains. (C and D) Close-up views of DUX4 (C) and HoxB1-Pbx1 heterodimer (D) bound to respective DNA substrate (as in A and B) highlighting distinct local interactions surrounding the key Trp residue docked in a hydrophobic pocket.
It is interesting that primates are distinct from the rest of mammals in having a negatively charged residue at position 70 (Figure 2D), which dictates the TAAT specificity for HD1. In all other DUXC family members, the amino acid at this position is arginine or glutamine, and indeed HD2 has an arginine at this position as does the DUX progenitor sDUX from marsupials, birds, and reptiles. Hendrickson et al. (2017) have shown that a subset of retroviral-like elements are regulated by DUX4 in early cleavage-stage embryos and that, in humans, these preferentially have the TAAT-containing putative DUX4 recognition site, whereas in mouse, the elements regulated by mDux preferentially have a TGAT-containing mDux recognition sequence. This suggests that the change from arginine to glutamic acid in DUX4 precipitated the coevolution of a cohort of mammalian endogenous retroviral elements throughout the human genome.
FSHD is caused by the cytotoxic effects of the DUX4 homeodomains recruiting p300 to target loci through the linked C terminus. Interestingly, both mDux and human DUX4 are toxic when the full-length proteins are overexpressed in mouse cells (Bosnakovski et al., 2009; Eidahl et al., 2016), and mDux is toxic when overexpressed in human cells (unpublished results). So, while there is apparently selective pressure on endogenous retroviral-like elements for optimal sequence specificity, this may not be the case with regard to the targets associated with cytotoxicity. On the other hand, B cell leukemic mutants of DUX4 invariably contain the homeodomains and lack the accompanying toxicity-associated transcriptional activation domain. The cancer phenotype of DUX4-IGH fusions is thus directly related to the DNA-binding spec-ificity of DUX4. It would be interesting to determine how leukemogenicity varies with sequence specificity alterations in HD1, and in particular whether an mDux mutant lacking its C terminus would be leukemogenic. Given the critical roles of the DUX4 double homeodomain in both FSHD and B cell leukemia, the structural basis of its target DNA sequence recognition presented here provides the framework for better understanding and potentially developing therapeutic strategies for these diseases.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Hideki Aihara (aihar001@umn.edu). Sharing of reagents may require MTA agreements.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Inducible C2C12 murine myoblasts (iC2C12) carrying DUX4 deletion constructs were cultured in DMEM, high glucose (HyClone Cat#SH30081.01), 1% penicillin/streptomycin (P/S) (Life Technologies #15140–122), 1% Glutamax (Life Technologies #SCR006), 1% Sodium Pyruvate (Caisson labs Cat#PYL01) and 20% FBS (Millipore #ESG1107, Temecula, CA), at 37°C in 5% CO2.
METHOD DETAILS
Structure determination
Human DUX4 (15–155) was expressed in E. coli strain BL21(DE3) with an N-terminal 6xHis-SUMO fusion tag and purified using nickel-affinity and size-exclusion chromatography. The SUMO-tag was removed by Ulp1 treatment during purification. The protein was mixed with a blunt-ended 17 bp double-stranded DNA substrate (5′-GCGTAATCTAATCAACA-3′ annealed to its complement) at 1:1.5 protein:DNA molar ratio in 20 mM Tris-HCl (pH 7.4), 150 mM sodium chloride, and 5 mM β-mercaptoethanol, and at an approximate protein concentration of 10 mg ml−1. We confirmed the formation of a homogeneous protein-DNA complex using size-exclusion chromatography. Crystals of the DUX4-(15–155)-DNA complex were grown by the hanging drop vapor diffusion method in a 24-well plate, using the reservoir solution consisting of 0.1 M BisTris-HCl (pH 6.5), 25%–30% polyethylene glycol 3,350, and 4%–10% glycerol. 1 μL each of the protein-DNA complex and the reservoir solutions were mixed to form the drops. The crystals were cryo-protected by increasing the glycerol concentration of the reservoir solutions to 25% then flash cooled by plunging into liquid nitrogen. X-ray diffraction data were collected at the NE-CAT (APS) beamlines 24-ID-C and 24-ID-E, and processed using XDS (Kabsch, 2010). The structure of the DUX4-DNA complex was solved by molecular replacement using the Pdx1 homeodomain (PDB ID: 2H1K) (Longo et al., 2007) as the search model in PHASER (McCoy et al., 2007). The atomic model was iteratively built using COOT (Emsley et al., 2010) and refined using PHENIX (Adams et al., 2010), imposing a standard set of protein geometry restrains as well as the base-pair and base-planarity restraints for DNA. Atomic displacement parameters refined included individual isotropic B-factors and a total of 8 TLS groups. A summary of data collection and model refinement statistics is shown in Table S1. Electron density suggested that Cys37 is covalently modified by β-mercaptoethanol, which is treated as ‘ligand’ in Table S1. Figures were generated using PyMOL (https://pymol.org/2/).
Generation of mutant cell lines & luciferase reporter assay
Terminal D4Z4 (2.7 kb) from p2lox-DUX4 was used as a template to generate all DUX4 mutation constructs (Bosnakovski et al., 2008b). Specific mutations were incorporated into PCR primers and amplified using LA Taq Polymerase (Takara BIO INC.). PCR fragments were fused together using In-Fusion HD Cloning Kit (Clontech) and cloned into p2lox plasmid. All of the constructs were sequenced before inserting into the targeting locus of iC2C12 myoblast cells as previously described (Bosnakovski et al., 2008b). Induction of every construct was confirmed by western blot using DUX4-specific antibody (R&D) and RTqPCR (Bosnakovski et al., 2008b). In addition, we confirmed by immunofluorescence that all of the mutant proteins exhibited nuclear localization. Cloning of the 2x DUX4 TAATCTAATCA luciferase reporter construct has been described previously (Zhang et al., 2016b). To generate the 2x TAATCTAATTA and TGATTCAATCA luciferase reporters, oligonucleotides encoding 2 copies of each potential DUX4 binding motif, but otherwise identical to each other and the original reporter, were synthesized and cloned into XhoI/HindIII-digested pGL4 lucif-erase reporter plasmid using In-Fusion HD Cloning Kit (Clontech). Positive clones were sequenced to confirm proper integration of the insert. For the luciferase assay, iC2C12-DUX4 and variant HD mutant cell lines were plated by flow cytometry using a FACS Aria into 96-well assay plates at a density of 2,000 cells/well. The following day, cells were transfected with pGL4-HD reporter plasmids (75 ng/well), using TransIT-LTI transfection reagent (Mirus Bio LLC). At 24 hours post-transfection, DUX4 expression was induced with 100 ng/mL doxycycline. Luciferase levels were quantified at 48 hours post-transfection using the ONE-Glo Luciferase Assay System (Promega), according to the manufacturer’s instructions. Luminescence was measured using the Cytation3 plate reader (BioTek). For each DUX4 construct, relative luciferase levels for the 3 different target sites are shown as normalized to the highest.
Electrophoretic mobility shift assay (EMSA)
5′-fluorescein-labeled 19 (18 for N2) nucleotide-long oligonucleotide annealed with an unlabeled complementary strand (Integrated DNA Technologies) at 15 nM was mixed with protein at indicated final concentrations in 15 mM Tris-HCl, pH 7.5, 75 mM NaCl, 3.7 mM tris(2-carboxyethyl)phosphine, 0.0075% Triton X-100, 0.1 mg/ml bovine serum albumin, 0.007% (w/v) bromophenol blue, and 10% (v/v) glycerol. The samples were separated on a non-denaturing 6% acrylamide gel (Invitrogen) with 0.5X TBE running buffer and the fluorescence was detected using a Typhoon FLA 9500 imager.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± SEM. Statistical analyses were done with Prism v6.07 (GraphPad Software, La Jolla, CA). The number of replicates and statistical method for each experiments were indicated in the corresponding figure legend.
DATA AND SOFTWARE AVAILABILITY
The atomic coordinates and structure factors for the DUX4-DNA complex crystal structure reported in this paper have been deposited in the Protein Data Bank, under the accession code 6E8C.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DUX4 | R&D Systems | Cat#MAB95351; RRID: AB_2754557 |
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) | Lucigen | Cat#60401–3 |
| E. coli 5α | New England BioLabs | Cat#C2987I |
| Stellar competent cells | Takara BIO INC. | Cat#636763 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM, high glucose | HyClone | Cat#SH30081.01 |
| FBS | PEAK serum | Cat#PS-FB3 |
| Penicillin/streptomycin | Life Technologies | Cat#15140–122 |
| Glutamax | Millipore | Cat#SCR006 |
| 0.25% Trypsin-EDTA | Life Technologies | Cat#25200–072 |
| PBS | HyClone | Cat#SH30256.01 |
| Doxycycline | Alfa Aesar | Cat#J60579–14 |
| XhoI | New England BioLabs | Cat#R0146S |
| HindIII-HF | New England BioLabs | Cat#R3104M |
| BsaI | New England BioLabs | Cat#R0535S |
| XbaI | New England BioLabs | Cat#R0145S |
| Recombinant Sumo protease (Ulp1 core) | This paper | N/A |
| LATaq Polymerase | Takara BIO INC. | Cat#RR02AG |
| In-Fusion HD Cloning Kit | Clontech | Cat#638909 |
| TransIT-LTI transfection reagent | Mirus Bio LLC | Cat#MIR2300 |
| QIAPrep Spin MiniPrep Kit | QIAGEN | Cat#27106 |
| Bis-Tris | Sigma | Cat#B9754–1KG |
| polyethylene glycol 3,350 | Hampton Research | Cat#HR2–591 |
| β-mercaptoethanol | Aldrich | Cat#M6250 |
| sodium chloride | Fisher | Cat#BP-358 |
| Triton X-100 | Acros | Cat#21568–2500 |
| Tris base | Fisher | Cat#BP152 |
| bovine serum albumin | Sigma | Cat#A7030 |
| bromophenol blue | Ricca Chemicals | Cat#1353–4 |
| glycerol | Fisher | Cat#S25342B |
| tris(2-carboxyethyl)phosphine-HCl | Biosynth | Cat#C-1818 |
| HisPur Ni-NTA resin | Thermo Scientific | Cat#88222 |
| Boric acid | Fisher | Cat#BP168–1 |
| EDTA | Fisher | Cat#BP120 |
| Imidazole | AK Scientific | Cat#D070 |
| Critical Commercial Assays | ||
| ONE-Glo Luciferase Assay System | Promega | Cat#E6120 |
| Deposited Data | ||
| Crystal structure data (atomic coordinates and structure factors) | This paper | PDB ID: 6E8C |
| Experimental Models: Cell Lines | ||
| iC2C12 | Bosnakovski et al., 2008b | N/A |
| Oligonucleotides | ||
| 5’-GCGTAATCTAATCAACA-3’ | Integrated DNA Technologies (IDT) | N/A |
| 5’-TGTTGATTAGATTACGC-3’ | IDT | N/A |
| /56-FAM/TGCGTAATCTAATCAACAC | IDT | N/A |
| GTGTTGATTAGATTACGCA | IDT | N/A |
| /56-FAM/TGCGTAATCTAATTAACAC | IDT | N/A |
| GTGTTAATTAGATTACGCA | IDT | N/A |
| /56-FAM/TGCGTGATCTAATCAACAC | IDT | N/A |
| GTGTTGATTAGATCACGCA | IDT | N/A |
| /56-FAM/TGCGTAATCTATCAACAC | IDT | N/A |
| GTGTTGATAGATTACGCA | IDT | N/A |
| /56-FAM/TT TCC CTT TTC CCC TTT TT | IDT | N/A |
| AAA AAG GGG AAA AGG GAA A | IDT | N/A |
| /56-FAM/AG CCC GCA CCA ACC ATG CC | IDT | N/A |
| GGC ATG GTT GGT GCG GGC T | IDT | N/A |
| /56-FAM/TG CGT AAT CTA GGG GAC AC | IDT | N/A |
| GTG TCC CCT AGA TTA CGC A | IDT | N/A |
| /56-FAM/TG CGC CCC CTA ATC AAC AC | IDT | N/A |
| GTG TTG ATT AGG GGG CGC A | IDT | N/A |
| Recombinant DNA | ||
| pE-SUMO-DUX4 (15–155) expression plasmid | This paper | N/A |
| p2lox | (Iacovino et al., 2011) | N/A |
| p2lox-DUX4 | (Bosnakovski et al., 2008b) | N/A |
| pGL4–2X-TAATCTAATCA | (Zhang et al., 2016b) | N/A |
| pGL4–2X-TAATCTAATTA | This paper | N/A |
| pGL4–2X-TGATTCAATCA | This paper | N/A |
| Software and Algorithms | ||
| XDS | (Kabsch, 2010) | N/A |
| PHASER | (McCoy et al., 2007) | N/A |
| COOT | (Emsley et al., 2010) | N/A |
| PHENIX | (Adams et al., 2010) | N/A |
| PyMOL | https://pymol.org/2/ | N/A |
| Prism (v6.07) | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| 6% acrylamide native gel (0.5x TBE) | Invitrogen | EC63652BOX |
| Superdex 200 26/60 | GE | 17-1071-01 |
Highlights.
DUX4 binds DNA with its tandem homeodomains (HDs) arranged head to head
DUX4 HD1 and HD2 bind different core sequences: TAAT and TGAT, respectively
A Glu70-Arg73 salt bridge in HD1 explains differential core sequence specificity
HD1-altered target specificity appears unique to primates
ACKNOWLEDGMENTS
We thank Cynthia S. Faraday for graphic design. This work was supported by grants from the Friends of FSH Research and NIH (R35 GM118047 to H.A.; R01 AR055685 to M.K.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by NIH (NIGMS P30-GM124165). The Pilatus 6M detector on 24-ID-C beam-line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.060.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aihara H, Shi K, Lee JK, Bosnakovski D, and Kyba M (2018). Comment on structural basis of DUX4/IGH-driven transactivation. Leukemia 32, 2090–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrane G, Soni A, and Ladias JA (2009). Structural basis for DNA recognition by the human PAX3 homeodomain. Biochemistry 48, 1148–1155. [DOI] [PubMed] [Google Scholar]
- Block GJ, Narayanan D, Amell AM, Petek LM, Davidson KC, Bird TD, Tawil R, Moon RT, and Miller DG (2013). Wnt/b-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet 22, 4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Lamb S, Simsek T, Xu Z, Belayew A, Perlingeiro R, and Kyba M (2008a). DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Exp. Neurol 214, 87–96. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppée F, et al. (2008b). An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 27, 2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Daughters RS, Xu Z, Slack JM, and Kyba M (2009). Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD-related repeats. PLoS One 4, e7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski D, Chan SSK, Recht OO, Hartweck LM, Gustafson CJ, Athman LL, Lowe DA, and Kyba M (2017). Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun 8, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, and Kyba M (2016). DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 44, 5161–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, and Hewitt JE (2007). Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet 81, 264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandapat A, Bosnakovski D, Hartweck LM, Arpke RW, Baltgalvis KA, Vang D, Baik J, Darabi R, Perlingeiro RC, Hamra FK, et al. (2014). Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep. 8, 1484–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zhang W, Wu H, Huang J, Zhang M, Wang P, Zhang H, Chen Z, Chen SJ, and Meng G (2018). Structural basis of DUX4/IGH-driven transactivation. Leukemia 32, 1466–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl JO, Giesige CR, Domire JS, Wallace LM, Fowler AM, Guckes SM, Garwick-Coppens SE, Labhart P, and Harper SQ (2016). Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum. Mol. Genet 25, 4577–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriëls J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, and Belayew A (1999). Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236, 25–32. [DOI] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, and Tapscott SJ (2012). DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JA, and Aggarwal AK (1995). Structure of the even-skipped home-odomain complexed to AT-rich DNA: new perspectives on homeodomain specificity. EMBO J. 14, 6280–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovino M, Bosnakovski D, Fey H, Rux D, Bajwa G, Mahen E, Mitanoska A, Xu Z, and Kyba M (2011). Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29, 1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R, Choudhury M, Zaman S, Singh S, Santosh V, Bastia D, and Escalante CR (2016). Functional architecture of the Reb1-Ter complex of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 113, E2267–E2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, and Pabo CO (1990). Crystal structure of an engrailed homeodomain-DNA complex at 2.8Å resolution: a framework for understanding homeodomain-DNA interactions. Cell 63, 579–590. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Rould MA, Aurora R, Herr W, and Pabo CO (1994). Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77, 21–32. [DOI] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Mattéotti C, Arias C, Corona ED, Nuñez NG, Leo O, et al. (2007). The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord 17, 611–623. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc NA, and Wolberger C (2003). Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17, 2060–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidenroth A, and Hewitt JE (2010). A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol. Biol 10, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. (2010). A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Stark MR, Johnson AD, and Wolberger C (1995). Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science 270, 262–269. [DOI] [PubMed] [Google Scholar]
- Longo A, Guanga GP, and Rose RB (2007). Structural basis for induced fit mechanisms in DNA recognition by the Pdx1 homeodomain. Biochemistry 46, 2948–2957. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Cryst 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passner JM, Ryoo HD, Shen L, Mann RS, and Aggarwal AK (1999). Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397, 714–719. [DOI] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, and Wolberger C (1999). Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96, 587–597. [DOI] [PubMed] [Google Scholar]
- Rickard AM, Petek LM, and Miller DG (2015). Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet 24, 5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, and Rohs R (2014). Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci 39, 381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, and Miller DG (2010). Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6, e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani VD, Punch VG, Kawabe Y, Jones AE, Palidwor GA, Porter CJ, Cross JW, Carvajal JJ, Kockx CE, van IJcken WF, et al. (2012). Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev. Cell 22, 1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, and Tapscott SJ (2017). Conservation and innovation in the DUX4-family gene network. Nat. Genet 49, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, et al. (1992). Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet 2, 26–30. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Guenther B, Desplan C, and Kuriyan J (1995). High resolution crystal structure of a Paired (Pax) class cooperative homeodomain dimer on DNA. Cell 82, 709–719. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, Imoto N, Kohsaka S, Kunita A, Doi K, et al. (2016). Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat. Genet 48, 569–574. [DOI] [PubMed] [Google Scholar]
- Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, Wu G, Li Y, Wei L, Iacobucci I, et al. ; St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome Project (2016a). Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet 48, 1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lee JK, Toso EA, Lee JS, Choi SH, Slattery M, Aihara H, and Kyba M (2016b). DNA-binding sequence specificity of DUX4. Skelet. Muscle 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for the DUX4-DNA complex crystal structure reported in this paper have been deposited in the Protein Data Bank, under the accession code 6E8C.