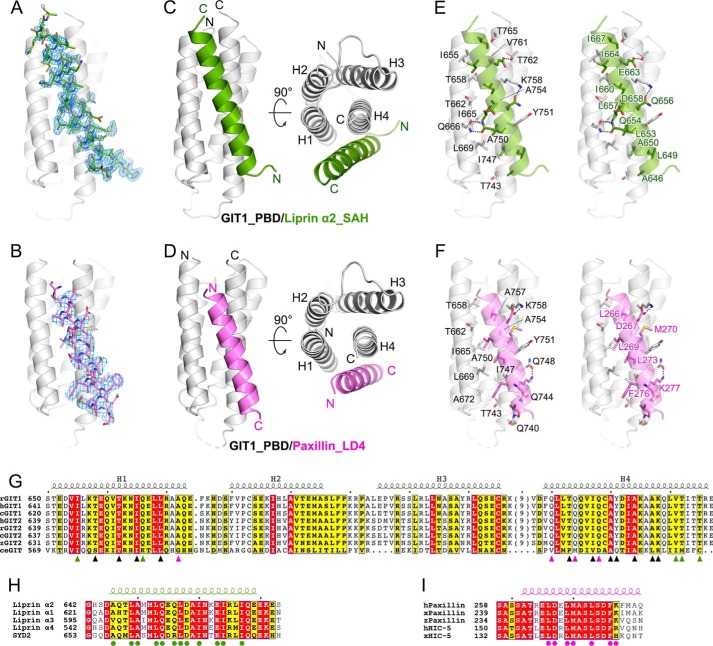
Figure 2.
The crystal structures of GIT1_PBD in complex with liprin-α2_SAH and paxillin_LD4. A and B, Fo − Fc omit maps (contoured at 2.5σ) of the bound SAH (A) and LD4 (B) peptides. The maps are shown with the final model of the corresponding complex superimposed. C and D, ribbon representations of the overall structures of the GIT1_PBD/liprin-α2_SAH (C) and GIT1_PBD/paxillin-LD4 (D) complexes. E and F, the atomic details of the GIT1_PBD/liprin-α2_SAH (E) and GIT1_PBD/paxillin-LD4 (F) interfaces. Hydrogen bonds and salt bridges are indicated by dashed lines. G, sequence alignment of the GIT family members from different species. Residues involved in SAH-specific, LD4-specific, and overlapped binding are marked with green, purple, and black triangles, respectively. The species are indicated as follows: human (h), rat (r), Xenopus tropicalis (x), for Danio rerio (z), Gallus gallus (c), and Caenorhabditis elegans (ce). The secondary structural elements are indicated above the alignment. H, sequence alignment of four liprin-α isoforms in humans and SYD2 in C. elegans. Residues involved in the GIT1_PBD/liprin-α_SAH interaction are marked with green circles. I, sequence alignment of paxillins and HIC-5s. Residues involved in the GIT1_PBD/paxillin-LD4 interaction are marked with purple circles.