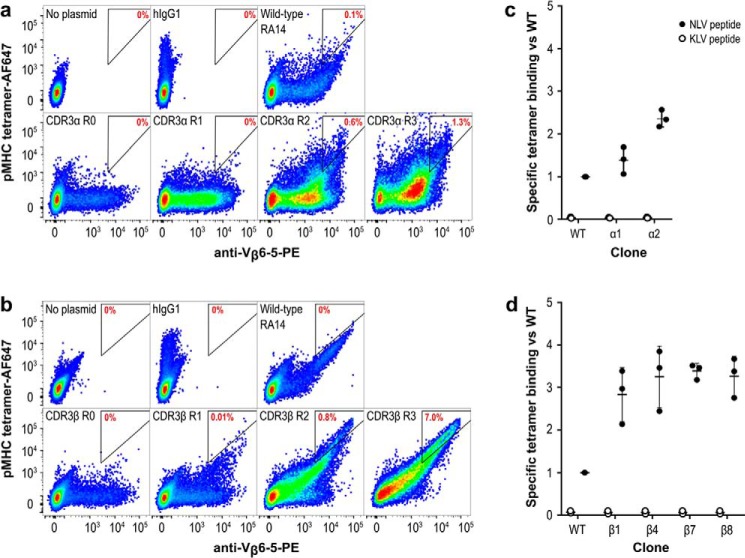
Figure 3.
RA14 variants with improved tetramer binding can be isolated by CHO display. The TCR display cassette optimized in Fig. 1 was mutagenized to create two libraries following the strategy in Fig. 2 and cloned into a pPy vector to allow for episomal maintenance in CHO-T cells. CDR3α (a) and CDR3β (b) libraries were separately transfected into CHO-T cells, stained, and sorted over three rounds to enrich for improved tetramer binding. Untransfected cells and cells displaying Fab hu1B7 (39) are shown as controls. The gate drawn is representative of the sorting gate used in round three, with the percentage of cells falling into the gate noted in red to facilitate comparisons. Individual clones selected during round three of the CDR3α (c) and CDR3β (d) libraries were re-transfected, stained, and assessed for specific tetramer binding relative to the WT RA14. Specific tetramer binding is the ratio of the AF647 signal (tetramer binding) to the PE signal (anti-TCRβ display) calculated on a per-cell basis. The median fluorescence intensity for this new variable was then normalized to the value for cells expressing WT RA14.