Abstract
GADD34 (growth arrest and DNA damage-inducible gene 34) plays a critical role in responses to DNA damage and endoplasmic reticulum stress. GADD34 has opposing effects on different stimuli-induced cell apoptosis events, but the reason for this is unclear. Here, using immunoblotting analyses and various molecular genetic approaches in HepG2 and SMMC-7721 cells, we demonstrate that GADD34 protects hepatocellular carcinoma (HCC) cells from tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)–induced apoptosis by stabilizing a BCL-2 family member, myeloid cell leukemia 1 (MCL-1). We found that GADD34 knockdown decreased MCL-1 levels and that GADD34 overexpression up-regulated MCL-1 expression in HCC cells. GADD34 did not affect MCL-1 transcription but enhanced MCL-1 protein stability. The proteasome inhibitor MG132 abrogated GADD34 depletion–induced MCL-1 down-regulation, suggesting that GADD34 inhibits the proteasomal degradation of MCL-1. Furthermore, GADD34 overexpression promoted extracellular signal-regulated kinase (ERK) phosphorylation through a signaling axis that consists of the E3 ubiquitin ligase tumor necrosis factor receptor–associated factor 6 (TRAF6) and transforming growth factor-β–activated kinase 1 (MAP3K7)–binding protein 1 (TAB1), which mediated the up-regulation of MCL-1 by GADD34. Of note, TRAIL up-regulated both GADD34 and MCL-1 levels, and knockdown of GADD34 and TRAF6 suppressed the induction of MCL-1 by TRAIL. Correspondingly, GADD34 knockdown potentiated TRAIL-induced apoptosis, and MCL-1 overexpression rescued TRAIL-treated and GADD34-depleted HCC cells from cell death. Taken together, these findings suggest that GADD34 inhibits TRAIL-induced HCC cell apoptosis through TRAF6- and ERK-mediated stabilization of MCL-1.
Keywords: apoptosis, cancer, drug resistance, oncogene, Trail, GADD34, MCL-1, TRAF
Introduction
GADD34 (growth arrest and DNA damage-inducible gene 34) plays a key role in DNA damage and endoplasmic reticulum (ER)2 stress response (1). Both DNA damage and ER stress can induce GADD34 expression, which leads to growth arrest. During ER stress response, eukaryotic initiation factor 2α (eIF2α) phosphorylation results in the inhibition of protein synthesis. Up-regulation of GADD34 in turn restores protein synthesis through recruiting protein phosphatase 1 to dephosphorylate eIF2α (2). Optimal stress response is required to help cells adapt to stressful insults, whereas persistent stress may lead to cell death. Previous studies indicate that GADD34 has opposing effects on apoptosis. Some studies show that GADD34 may induce apoptosis (3, 4), whereas other studies demonstrate that GADD34 can prevent apoptosis or tissue injury (5–8). The mechanisms underlying the anti-apoptotic effect of GADD34 remain elusive. We hypothesized that GADD34 may regulate prosurvival genes in residual cancer cells that are treated by anti-cancer agents.
BCL-2, BCL-xL, and myeloid cell leukemia-1 (MCL-1) are three BCL-2 family proteins that have key roles in apoptosis (9). MCL-1 stabilizes mitochondrial membrane and inhibits the release of cytochrome C. Meanwhile, MCL-1 heterodimerizes with the pro-apoptosis members in BCL-2 family, such as Bim and Bak, thereby inhibiting the pro-apoptic effects of Bim/Bak (10). Thus, MCL-1 plays key roles in inhibiting apoptosis. MCL-1 is a key survival factor in many cancers (11). MCL-1 overexpression inhibits cell death and promotes tumorigenesis. Moreover, MCL-1 is involved in chemotherapy resistance and tumor recurrence (12). The expression of MCL-1 can be promoted by multiple signaling pathways, such as JAK/STAT and PI3K/Akt pathways (13, 14). In addition, post-translational modifications of the PEST domains in MCL-1 regulate the stability of MCL-1 protein (15). MULE and β-Trcp are two ubiquitin ligases that mediate ubiquitin-proteasomal degradation of MCL-1 (16, 17). Inhibition of MCL-1 by oligonucleotide, BH-3 domain mimetic, and small molecule inhibitors is an attractive strategy to treat cancer (18). So far, it is unknown whether GADD34 may regulate apoptosis through BCL-2 family.
Tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) specifically induces apoptosis in tumor instead of normal cells. TRAIL can initiate cancer cell apoptosis by binding to death receptor 4 (DR4) and DR5 (19). Therefore, biotherapeutic death receptors agonists hold promise in cancer therapy. Indeed, TRAIL can induce cancer cell apoptosis without causing toxicity in preclinical mouse models (20). However, the outcome of clinical trials with death receptors agonists has been disappointing so far. Notably, the sensitivity of cancer cells to TRAIL-induced apoptosis may vary among different types of cancer. Hepatoma cells are relatively insensitive to TRAIL. Moreover, there is endogenous TRAIL in many cancers. Increased expression of endogenous TRAIL is associated with poor prognosis in cancers that are resistant to TRAIL-induced apoptosis (20). Therefore, it is important to better understand the biology of TRAIL-death receptor signaling and the mechanisms underlying TRAIL resistance to meet the challenges for effectively targeting this pathway (21).
In this study, we demonstrate that GADD34 positively regulates MCL-1 expression in HCC cells. GADD34 up-regulates MCL-1 expression through suppressing proteasomal degradation of MCL-1 protein. Mechanistically, the stabilization of MCL-1 by GADD34 is mediated by extracellular signal-regulated kinase (ERK). GADD34 promotes ERK1/2 phosphorylation via tumor necrosis factor receptor–associated factor 6 (TRAF6) and transforming growth factor-β–activated kinase 1 (MAP3K7)-binding protein 1 (TAB1). Knockdown of GADD34 enhances TRAIL-induced apoptosis in HCC cells. Finally, overexpression of MCL-1 abrogates the potentiation of TRAIL-induced apoptosis by GADD34 depletion.
Results
GADD34 up-regulates MCL-1 expression in HCC cells
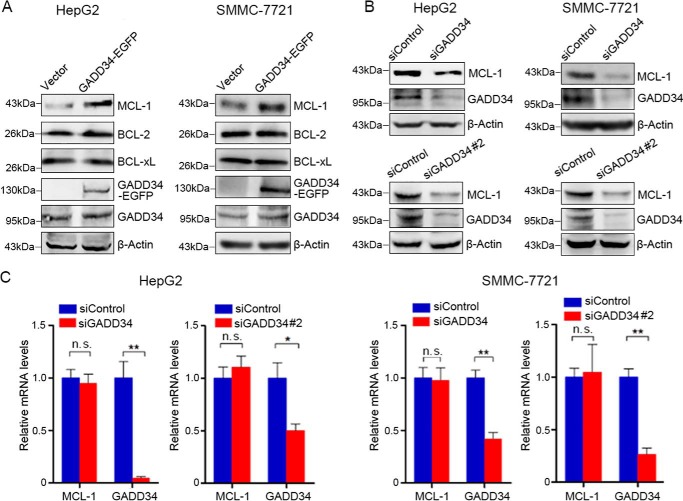
To determine the effects of GADD34 on BCL-2 family proteins, EGFP-tagged GADD34 was overexpressed in HepG2 cells, followed by Western blotting analysis of BCL-2, BCL-xL, and MCL-1 expression. Overexpression of GADD34 resulted in an increase in the levels of MCL-1, although it had no effect on BCL-2 and BCL-xL expression (Fig. 1A). Similar effects were detected in SMMC-7721 (Fig. 1A). Knockdown of GADD34 by two siRNA consistently resulted in decreased MCL-1 levels in both HepG2 and SMMC-7721 cells (Fig. 1B). In addition, GADD34 up-regulated MCL-1 expression in Hep3B cells (Fig. S1). Together, these results demonstrate that GADD34 up-regulates MCL-1 in HCC cells. To determine whether GADD34 affects MCL-1 transcription, HepG2 and SMMC-7721 cells were transfected with control or GADD34 siRNA, followed by real-time RT-PCR analysis of MCL-1 transcription. GADD34 knockdown had no effect on the transcription levels of MCL-1 (Fig. 1C).
Figure 1.
GADD34 up-regulates MCL-1 protein expression in HCC cells. A, HepG2 and SMMC-7721 cells were transfected with the EGFP-tagged GADD34 expression plasmid or EGFP vector, followed by Western blotting analysis of MCL-1, BCL-2, BCL-xL, and GADD34 expression. B, HepG2 and SMMC-7721 cells were transfected with siControl, siGADD34, or siGADD34#2, followed by Western blotting analysis of MCL-1 and GADD34 expression. C, HepG2 and SMMC-7721 cells were transfected with siControl, siGADD34, or siGADD34#2, followed by real-time RT-PCR analysis of MCL-1 and GADD34 transcription. The relative levels of MCL-1 and GADD34 mRNA were plotted. The values represent the means ± S.D. (n = 3). **, p < 0.01.
GADD34 suppresses proteasomal degradation of MCL-1
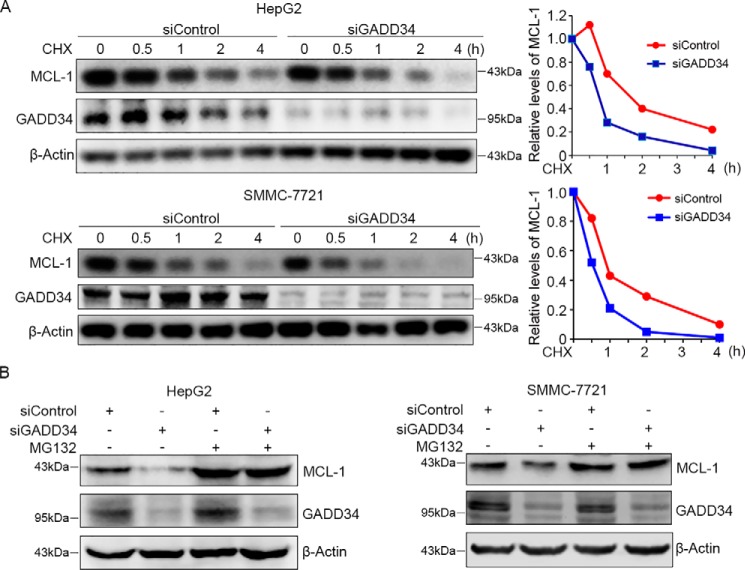
To determine whether GADD34 affects the stability of MCL-1, HepG2 cells were transfected with control or GADD34 siRNA, followed by treatment with the protein synthesis inhibitor cycloheximide (CHX) and detection of MCL-1 levels at different periods. While the levels of MCL-1 dropped gradually after treatment with CHX, GADD34 knockdown further accelerated MCL-1 protein turnover in HepG2 cells (Fig. 2A). Similar effects were detected in SMMC-7721 cells (Fig. 2A).
Figure 2.
GADD34 knockdown accelerates MCL-1 protein degradation. A, HepG2 and SMMC-7721 cells were transfected with siControl or siGADD34, followed by treatment with 100 μg/ml of CHX for indicated times, and Western blotting analysis of MCL-1 and GADD34. The relative levels of MCL-1 were plotted. B, HepG2 and SMMC-7721 cells were transfected with siControl or siGADD34 and treated with or without 5 μm of MG132 for 24 h, followed by Western blotting analysis of MCL-1 and GADD34.
To determine whether GADD34 inhibits the proteasomal degradation of MCL-1, HepG2 and SMMC-7721 cells were transfected with control or GADD34 siRNA, followed by treatment with or without proteasome inhibitor MG132 and Western blotting analysis of MCL-1. Treatment with MG132 led to an increase in MCL-1 levels and abrogated the down-regulation of MCL-1 by GADD34 depletion in both HepG2 and SMMC-7721 cells (Fig. 2B). Together, these data demonstrate that GADD34 suppresses proteasomal degradation of MCL-1.
GADD34 up-regulates MCL-1 by promoting ERK1/2 phosphorylation
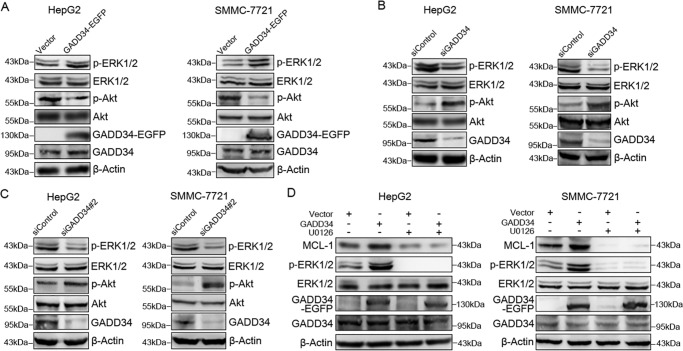
To determine the mechanism underlying the stabilization of MCL-1 by GADD34, we detected whether GADD34 physically interacted with MCL-1 protein. Immunoprecipitation assay demonstrated no interaction between GADD34 and MCL-1 (data not shown), suggesting that GADD34 may indirectly regulate MCL-1. Previous reports indicated that ERK1/2 could suppress MCL-1 degradation by phosphorylating PEST domains in MCL-1 (15). To detect whether GADD34 affects ERK1/2 phosphorylation, GADD34 was overexpressed in HepG2 cells, followed by Western blotting analysis of ERK1/2 phosphorylation. Overexpression of GADD34 promoted ERK1/2 phosphorylation in HepG2 cells (Fig. 3A). Consistent with previous studies that showed GADD34 negatively regulated Akt phosphorylation, overexpression of GADD34 inhibited Akt phosphorylation (Fig. 3A). GADD34 overexpression also promoted ERK1/2 phosphorylation and inhibited Akt phosphorylation in SMMC-7721 and Hep3B cells (Fig. 3A and Fig. S1A).
Figure 3.
GADD34 up-regulates MCL-1 through TRAF6 and ERK1/2. A, HepG2 and SMMC-7721 cells were transfected with the EGFP-tagged GADD34 expression plasmid or EGFP vector, followed by Western blotting analysis of p-ERK1/2 (Thr202/Tyr204), ERK1/2, p-Akt (Ser473), Akt, and GADD34. B, HepG2 and SMMC-7721 cells were transfected with siControl or siGADD34, followed by Western blotting analysis of p-ERK1/2 (Thr202/Tyr204), ERK1/2, p-Akt (Ser473), Akt, and GADD34. C, HepG2 and SMMC-7721 cells were transfected with siControl or siGADD34#2, followed by Western blotting analysis of p-ERK1/2 (Thr202/Tyr204), ERK1/2, p-Akt (Ser473), Akt, and GADD34. D, HepG2 and SMMC-7721 cells were transfected with the EGFP-tagged GADD34 expression plasmid or EGFP vector and treated with or without 10 μm of U0126 for 48 h, followed by Western blotting analysis of MCL-1, p-ERK1/2 (Thr202/Tyr204), ERK1/2, and GADD34.
In addition, we transfected HepG2 and SMMC-7721 cells with control or GADD34 siRNA, followed by Western blotting analysis of ERK1/2 and Akt phosphorylation. GADD34 knockdown by two siRNA consistently led to a decrease in ERK1/2 phosphorylation and an increase in Akt phosphorylation in both HepG2 and SMMC-7721 cells (Fig. 3, B and C). Similar effects were detected in Hep3B cells (Fig. S1B). Although overexpression of GADD34 increased ERK1/2 phosphorylation and MCL-1 expression, treatment with the MEK inhibitor U0126 antagonized the induction of ERK1/2 phosphorylation and MCL-1 expression by GADD34 (Fig. 3D), indicating that the MEK-ERK1/2 pathway mediates the up-regulation of MCL-1 by GADD34.
TRAF6 mediates the up-regulation of ERK1/2 phosphorylation and MCL-1 expression by GADD34
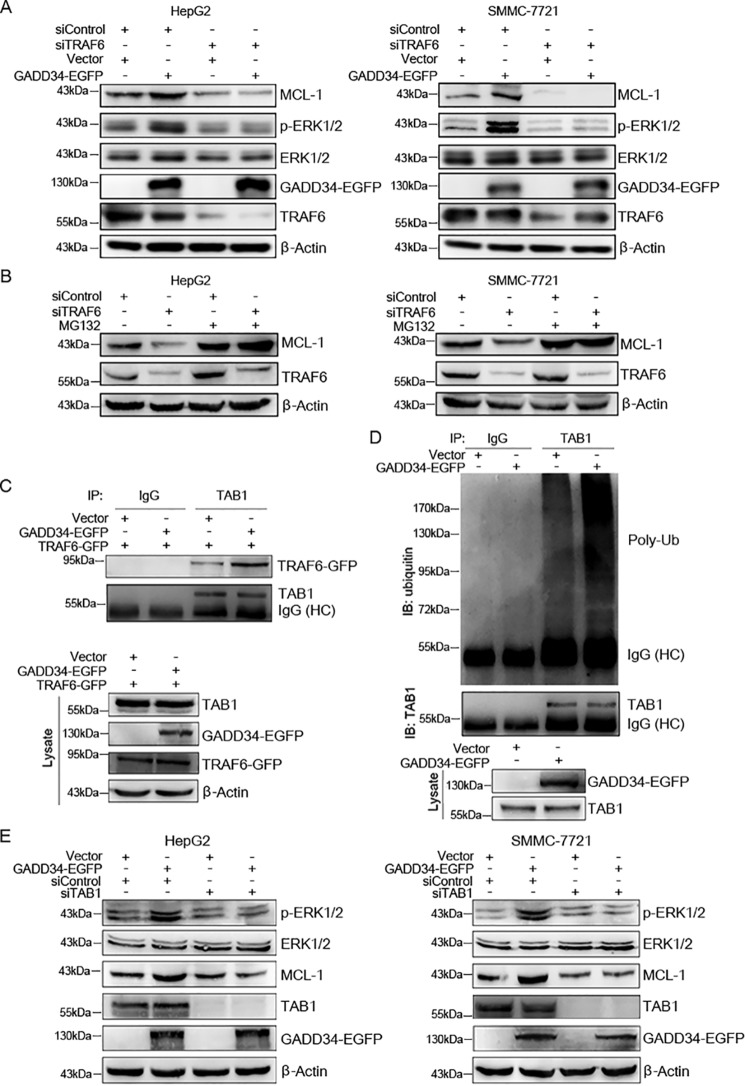
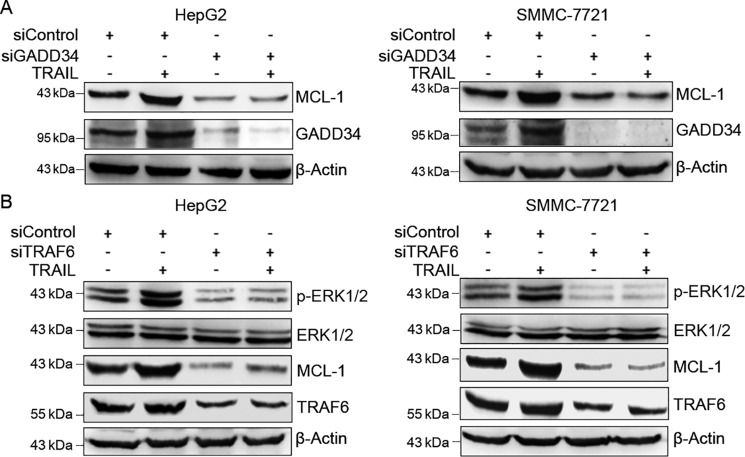
Previous studies show that GADD34 binds to the E3 ubiquitin ligase TRAF6 (22). While TRAF6 mediates the ubiquitination of Akt and subsequently promotes Akt phosphorylation, it stimulates ERK1/2 phosphorylation in Ras-independent manner (22, 23). To determine whether TRAF6 is involved in the regulation of ERK1/2 phosphorylation and MCL-1 expression by GADD34, HepG2 cells were transfected with or without EGFP-tagged GADD34 expression plasmid, followed by transfection with control siRNA or TRAF6 siRNA, and Western blotting analysis of ERK1/2 phosphorylation and MCL-1 expression. TRAF6 knockdown abrogated the up-regulation of ERK1/2 phosphorylation and MCL-1 expression by GADD34 in HepG2 cells (Fig. 4A). Similar results were detected in SMMC-7721 cells (Fig. 4A). Together, these data indicate that TRAF6 contributes, at least in part, to the up-regulation of MCL-1 expression by GADD34.
Figure 4.
TRAF6 and TAB1 mediate the up-regulation of ERK1/2 phosphorylation and MCL-1 expression by GADD34. A, HepG2 and SMMC-7721 cells were transfected with the EGFP-tagged GADD34 expression plasmid or EGFP vector and transfected with siControl or siTRAF6, followed by Western blotting analysis of MCL-1, p-ERK1/2 (Thr202/Tyr204), ERK1/2, TRAF6, and GADD34. B, HepG2 and SMMC-7721 cells were transfected with siControl or siTRAF6 and treated with or without 5 μm of MG132 for 24 h, followed by Western blotting analysis of MCL-1 and TRAF6. C, HepG2 cells were transfected with or without EGFP-tagged GADD34 and GFP-tagged TRAF6 expression plasmid, followed by immunoprecipitation with anti-TAB1 antibody and Western blotting analysis of TAB1 and TRAF6. The levels of GADD34–EGFP, TRAF6–GFP, and TAB1 in whole cell lysates were also detected by Western blotting. D, HepG2 cells were transfected with the EGFP-tagged GADD34 expression plasmid or EGFP vector, followed by immunoprecipitation with anti-TAB1 antibody and Western blotting analysis of TAB1 polyubiquitination (Poly-Ub). The levels of TAB1 and GADD34–EGFP in whole cell lysates were detected by Western blotting. E, HepG2 and SMMC-7721 cells were transfected with the EGFP-tagged GADD34 expression plasmid or EGFP vector and transfected with siControl or siTAB1, followed by Western blotting analysis of MCL-1, p-ERK1/2 (Thr202/Tyr204), ERK1/2, TAB1, and GADD34. IB, immunoblotting; IP, immunoprecipitation.
Because ERK1/2 could phosphorylate and stabilize MCL-1 protein, we then detected whether TRAF6 prevents proteasomal degradation of MCL-1. HepG2 cells were transfected with or without control siRNA or TRAF6 siRNA, followed by treatment with or without proteasome inhibitor MG132 and Western blotting analysis of MCL-1. MG132 abrogated the down-regulation of MCL-1 by TRAF6 knockdown in both HepG2 and SMMC-7721 cells (Fig. 4B). Together, these data demonstrate that TRAF6 cooperates with GADD34 to suppress proteasomal degradation of MCL-1.
Previous studies demonstrate that TRAF6 interacts with TAB1 and induces nondegradative ubiquitination of TAB1, thereby activating TAB1 (24). Ubiquitinated TAB can activate MAPKs such as p38 and ERK (25). To determine whether GADD34 regulates the interaction between TRAF6 and TAB1, GADD34 was overexpressed in HepG2 cells, followed by immunoprecipitation of TAB1. Because the immunoprecipitated TRAF6 protein is poorly separated from the heavy chain of IgG in Western blotting, GFP-tagged TRAF6 was transfected into HepG2 cells, and the interaction between TAB1 and TRAF6–GFP was detected. Overexpression of GADD34 enhanced the interaction between TRAF6 and TAB1 (Fig. 4C). Meanwhile, overexpression of GADD34 promoted TAB1 ubiquitination, whereas it did not affect the levels of TAB1 protein (Fig. 4D). Furthermore, TAB1 knockdown abrogated the up-regulation of ERK1/2 phosphorylation and MCL-1 expression by GADD34 (Fig. 4E). Taken together, these data suggest that GADD34 promotes ERK1/2 phosphorylation and MCL-1 expression through the TRAF6–TAB1 axis.
GADD34 knockdown inhibits the up-regulation of MCL-1 by TRAIL
To detect the effect of TRAIL on GADD34 and MCL-1 expression in HCC cells, we treated HepG2 and SMMC-7721 cells with TRAIL for 24 h, followed by Western blotting analysis of GADD34 and MCL-1 expression. Treatment of HepG2 cells with TRAIL moderately induced MCL-1 and GADD34 expression in both HepG2 and SMMC-7721 cells (Fig. 5A). In addition, GADD34 knockdown down-regulated MCL-1 levels and abrogated the induction of MCL-1 by TRAIL in both HepG2 and SMMC-7721 cells (Fig. 5A), indicating that GADD34 mediates the up-regulation of MCL-1 by TRAIL.
Figure 5.
GADD34 and TRAF6 mediate the up-regulation of MCL-1 by TRAIL. A, HepG2 and SMMC-7721 cells were transfected with siControl or siGADD34, followed by treatment with 100 ng/ml of TRAIL for 24 h, and Western blotting analysis of MCL-1 and GADD34. B, HepG2 and SMMC-7721 cells were transfected with siControl or siTRAF6, followed by treatment with or without 100 ng/ml of TRAIL for 24 h and Western blotting analysis of MCL-1, p-ERK1/2 (Thr202/Tyr204), ERK1/2, and TRAF6.
Given that TRAF6 and ERK1/2 mediate the stabilization of MCL-1 by GADD34, we then detected the effect of TRAF6 knockdown on TRAIL-induced ERK1/2 phosphorylation and MCL-1 expression. TRAF6 knockdown resulted in a decrease in ERK1/2 phosphorylation and MCL-1 expression in both HepG2 and SMMC-7721 cells (Fig. 5B). In addition, TRAF6 knockdown abrogated the induction of ERK1/2 phosphorylation and MCL-1 expression by TRAIL (Fig. 5B).
GADD34 knockdown potentiates TRAIL-induced apoptosis in HCC cells
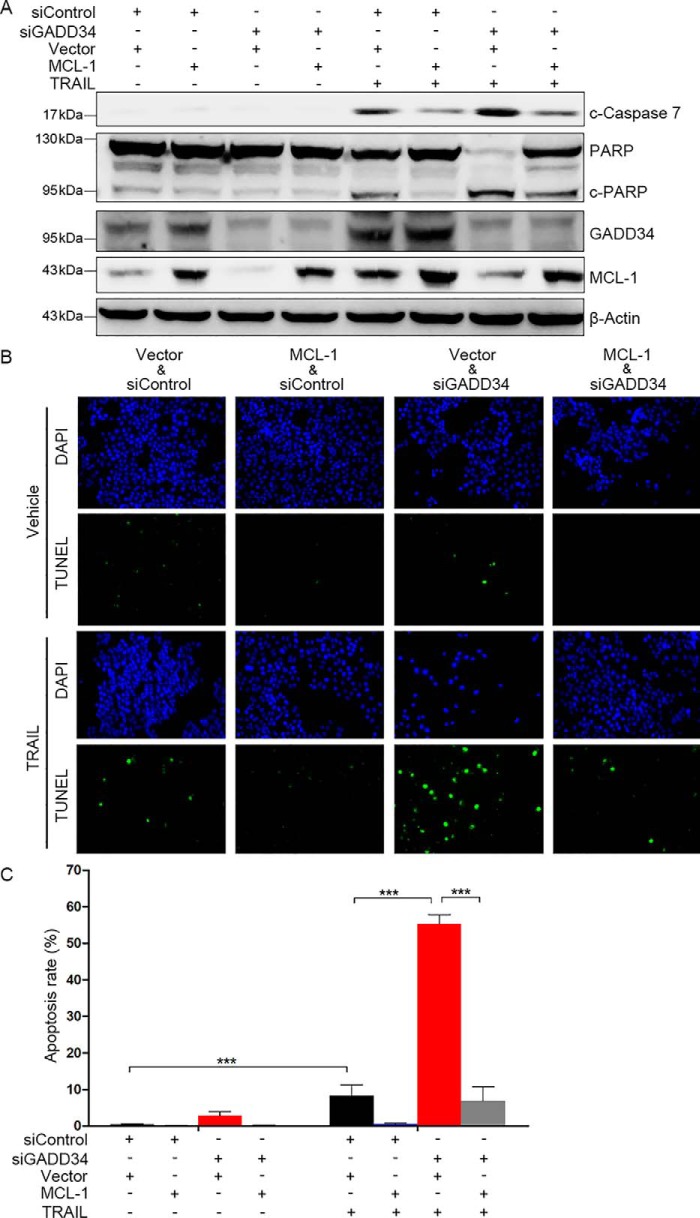
TRAIL can induce caspase-dependent apoptosis. To determine the effects of GADD34 and MCL-1 on TRAIL-induced cleavage of the effector caspase 7 and PARP, HepG2 cells were transfected with MCL-1 plasmid or the empty vector, followed by transfection of siControl or siGADD34 and treatment with or without TRAIL. GADD34 knockdown led to increased cleavage of caspase 7 and PARP in HepG2 cells (Fig. 6A), indicating that GADD34 inhibited caspase activation. Overexpression of MCL-1 inhibited TRAIL-induced cleavage of caspase 7 and PARP and suppressed the potentiation of TRAIL-induced caspase 7 and PARP cleavage by GADD34 depletion (Fig. 6A).
Figure 6.
MCL-1 suppresses the potentiation of TRAIL-induced apoptosis by GADD34 depletion in HepG2 cells. A, HepG2 cells were transfected with siControl or siGADD34, and the MCL-1 expression plasmid or vector, followed by treatment with or without 100 ng/ml of TRAIL for 24 h. The levels of cleaved caspase-7, PARP, GADD34, and MCL-1 were detected by Western blotting. B, HepG2 cells were transfected with siControl or siGADD34 and the MCL-1 expression plasmid or vector followed by treatment with or without 100 ng/ml of TRAIL for 24 h. Cell apoptosis was detected by TUNEL assays. C, the apoptotic rate was plotted. The values represent the means ± S.D. (n = 3). ***, p < 0.001. DAPI, 4′,6′-diamino-2-phenylindole.
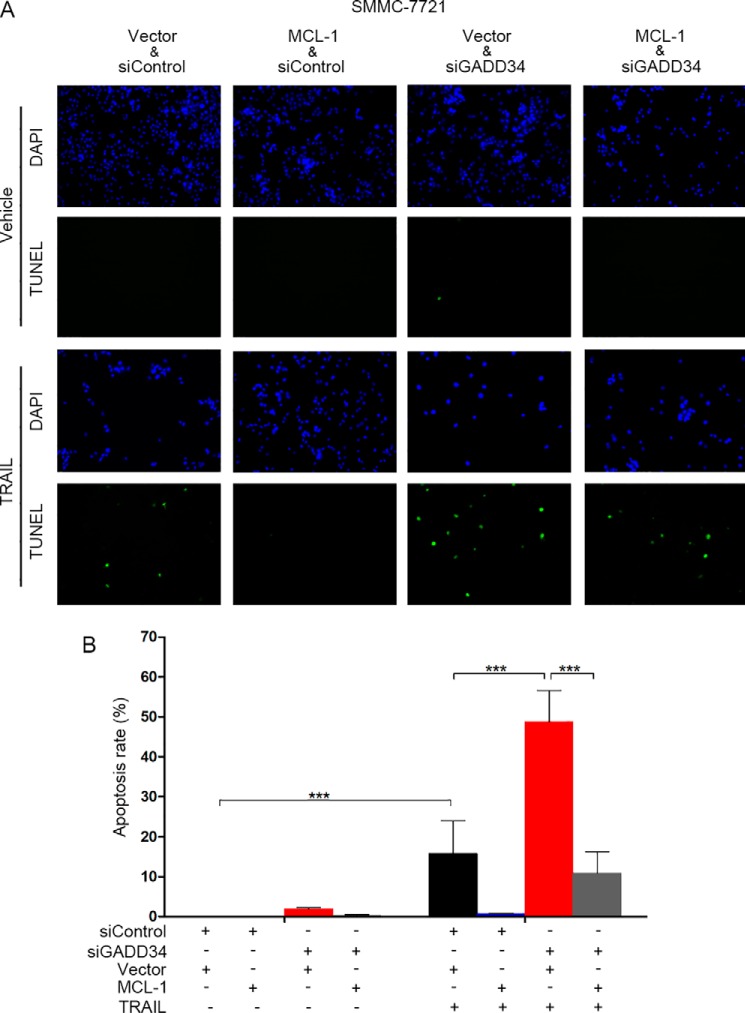
Next, we investigated the effects of GADD34 and MCL-1 on TRAIL-induced apoptosis. HepG2 cells were transfected with MCL-1 plasmid or the empty vector, followed by transfection of siControl or siGADD34 and treatment with or without TRAIL. TUNEL assays demonstrated that knockdown of GADD34 led to an increase in TRAIL-induced apoptosis (Fig. 6, B and C). Furthermore, overexpression of MCL-1 inhibited TRAIL-induced apoptosis and compromised the potentiation of TRAIL-induced apoptosis by GADD34 knockdown (Fig. 6, B and C). Similar effects were detected in SMMC-7721 cells (Fig. 7). Together, these data indicate that MCL-1 mediates, at least in part, the prevention of TRAIL-induced apoptosis by GADD34. To determine the effect of GADD34 on other chemotherapeutic agent-induced apoptosis, we detected the effect of GADD34 on Taxol-induced apoptosis. GADD34 knockdown led to an increase in Taxol-induced apoptosis in both HepG2 and SMMC-7721 cells (Fig. S2).
Figure 7.
Overexpression of MCL-1 rescues TRAIL-treated and GADD34-depleted SMMC-7721 cells from apoptosis. A, SMMC-7721 cells were transfected with siControl or siGADD34 and the MCL-1 expression plasmid or vector followed by treatment with or without 100 ng/ml of TRAIL for 24 h. Cell apoptosis was detected by TUNEL assays. B, the apoptotic rate was plotted. The values represent the means ± S.D. (n = 3). ***, p < 0.001. DAPI, 4′,6′-diamino-2-phenylindole.
Discussion
In this study, the mechanisms underlying the prevention of TRAIL-induced apoptosis by GADD34 were investigated. Previous studies have demonstrated that recombinant TRAIL may be a promising anti-cancer agent that selectively induces cancer cell apoptosis (20). In addition, TRAIL can be induced by some synthetic or natural agents, thereby mediating the anticancer effects of these agents through activating TRAIL apoptosis pathway (26). However, accumulating evidence show that TRAIL also promotes prosurvival, proliferative, or migratory signaling, such as NF-κB, PI3K/Akt, and MAPK signaling, which lead to TRAIL resistance in cancer cells (27). Similar to other anticancer agents, TRAIL resistance is a critical problem to compromise its anticancer effect. The mechanisms underlying TRAIL resistance in cancer therapy may be complex. Whereas both DR4 and DR5 mediate the pro-apoptosis effect of TRAIL, the decoy death receptors may be overexpressed in some cancer cells, which leads to neutralization of TRAIL. Therefore, the levels of death receptors and decoy receptors in cancer cells may affect TRAIL sensitivity. In addition, c-FLIP (cellular FLICE (FADD-like interleukin-1β–converting enzyme)–inhibitory protein) is involved in the resistance of cancer cells to TRAIL-induced cell death (28, 29). Levels of c-FLIP may affect the sensitivity of cancer cells to TRAIL (30). In the current study, we show that GADD34 prevents TRAIL-induced apoptosis through TRAF6- and ERK-mediated stabilization of the BCL-2 family member MCL-1.
GADD34 can be induced by DNA damage, hypoxia, and endoplasmic reticulum stress. Early studies demonstrate that GADD34 promotes ion radiation- and proteasome inhibitor-induced apoptosis (31, 32). Thus, GADD34 is generally considered as a tumor suppressor. In contrast, GADD34 knockdown enhances palmitate-induced mouse insulinoma cell apoptosis (7). Moreover, GADD34 knockout results in more liver cell apoptosis in lipopolysaccharide-treated mice (5). GADD34 inhibits macrophages apoptosis induced by lipopolysaccharide in combination with amino acid deprivation (6). Our current study demonstrates that GADD34 inhibits TRAIL- and Taxol-induced apoptosis. Therefore, GADD34 may regulate cell apoptosis in context-dependent manner. Similar to other sensors of cell stress, such as p53, GADD34 may be cytoprotective and cytotoxic, depending on the type or degree of damage. Furthermore, GADD34 reportedly promotes tumor growth by evasion of immune surveillance (33). In addition, GADD34 up-regulates the expression of pro-inflammatory cytokines and thereby promotes azoxymethane/dextran sodium sulfate-induced colorectal carcinogenesis (34). Key apoptotic proteins as FADD and procaspase-8 were also involved in transducing nonapoptotic signaling in response to TRAIL (27).
It is well-documented that MCL-1 is less stable than other BCL-2 family members because of the presence of PEST sequences at its N terminus. ERK directly phosphorylates MCL-1 at both Thr163 and Thr92, whereas Thr163 is the major phosphorylation site. Phosphorylation of MCL-1 by ERK leads to increased interaction between MCL-1 and Pin1, which prevents MCL-1 degradation (35). Our current study demonstrates that GADD34 prevents TRAIL-induced apoptosis. Mechanistically, GADD34 up-regulates ERK1/2 phosphorylation in TRAF6-dependent manner, which in turn stabilizes MCL-1. Notably, TRAF6 can promote ERK1/2 phosphorylation independent of Ras (23, 36, 37). Although GADD34 suppresses TRAF6-mediated Akt activation (22), the current study demonstrates that GADD34 promotes the interaction between TRAF6 and TAB1, leading to increased TAB1 ubiquitination, ERK phosphorylation, and MCL-1 stabilization. Therefore, GADD34 may act as a switch between PI3K/Akt and MAPK signaling pathways. As an anti-apoptotic protein, MCL-1 contributes to cell survival, which makes it an ideal target of anticancer drugs. The MCL-1 inhibitor S63845 potently inhibits tumor growth by activating BAX/BAK-dependent mitochondrial apoptotic pathway (38). Previous studies have demonstrated that down-regulation of MCL-1 can sensitize cancer cells to TRAIL and other chemotherapeutic agents (39–41). The current study demonstrates that GADD34 is a positive regulator of MCL-1 and a negative regulator of TRAIL- and Taxol-induced apoptosis.
Anticancer agents may inevitably induce stress response in cancer cells. A primary function of stress response is to adapt to exogenous insults and survive at the expense of growth arrest. In fact, growth arrest and survival is a mechanism for cancer cells to resist proteasome inhibitor therapy (42). The same may be true for TRAIL resistance. Given that GADD34 can induce both growth arrest and survival, which may promote cancer cells entering a state of cellular dormancy and help dormant cells remain viable in a quiescent state. Cancer cell dormancy is one of major cause of cancer recurrence and drug resistance (43). We speculate that the induction of GADD34 by TRAIL may be a survival/adaptation response. Upon withdrawal of TRAIL, GADD34 expression may restore to a lower level, which allows cancer cells exit from dormancy. Mechanistically, GADD34 may promote TRAIL resistance by dephosphorylating eIF2α and stabilizing MCL-1. Our previous study demonstrated that inhibition of eIF2α dephosphorylation could sensitize HCC cells to TRAIL (8). The current study provides a new mechanism for the prosurvival function of GADD34 in HCC cells. Thus, in addition to the inhibition of eIF2α dephosphorylation, inhibition of MCL-1 may be another strategy to prevent GADD34 from facilitating adaption to TRAIL treatment or endogenous TRAIL.
Experimental procedures
Cell culture
HCC cell lines HepG2, SMMC-7721, and Hep3B were obtained from Cell Lines Bank, Chinese Academy of Science (Shanghai, China). The cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% new born calf serum (Thermo Fisher Scientific, Waltham, MA). The cells were incubated at 37 °C in a humidified atmosphere of 5% CO2.
Reagents and antibodies
Recombinant human TRAIL/Apo2L was purchased from Pepro Tech Inc. (Rocky Hill, NJ). MG132 and Taxol were from Merck–Millipore. U0126 and CHX were from Beyotime Biotechnology (Jiangsu, China). Protein A/G magnetic beads were from Bimake (Selleck Chemical, Houston, TX). The deubiquitinase inhibitor PR-619 was from TargetMol (Shanghai, China). The antibodies used were as follows: anti-MCL-1, anti-p-ERK1/2, anti-ERK1/2, anti-Akt, anti-BCL-2, and anti-BCL-xL antibodies were purchased from Cell Signaling Technology (Beverly, MA); anti-p-Akt was from Epitomics (Burlingame, CA); anti-GADD34 antibody was purchased from Gene Tex Inc. (Irvine, CA); anti-TRAF6 and anti-TAB1 antibodies were purchased from Proteintech (Rosemont, IL); and anti-ubiquitin and anti-β-actin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
RNAi
All siRNAs were custom-synthesized products of GenePharma Co. Ltd. (Shanghai, China). The two target sequences for GADD34 knockdown are as follows: 5′-GGACACUGCAAGGUUCUGA-3′ for siGADD34 and 5′-GUCAAUUUGCAGAUGGCCATT-3′ for siGADD34#2. The target sequences of MCL-1, TRAF6, and TAB1 siRNA are 5′-AAGUAUCACAGACGUUCUC-3′, 5′-GGAAACUAUUCACCAGUUATT-3′, and 5′-CUGCGAUGAUUGACACUGA-3′, respectively. Proliferating cells were incubated with 50 nm siRNA in 2 ml of OPTI-MEM I reduced serum medium containing Lipofectamine RNAiMAX (Thermo Fisher Scientific). 48 h after transfections, the cells were harvested for further experiments.
Plasmid construction
The plasmid for GADD34–EGFP was purchased from Bio-Atom Biotechnology (Chengdu, China). MCL-1 plasmid was generated by subcloning human MCL-1 cDNA into pTango-zeo (Bio-Atom Biotechnology, Chengdu, China). TRAF6–GFP plasmid was purchased from Vigene Biosciences (Rockville, MD). For overexpression of MCL-1 or TRAF6 in cells, MCL-1 or TRAF6 plasmid was preincubated with Lipofectamine 2000 and added into the culture medium with a final concentration of 2 μg/ml.
Western blotting
The total cellular samples were washed twice with ice-cold PBS and lysed in ice-cold lysis buffer (1% Triton X-100, 40 mm Hepes, pH 7.5, 120 mm NaCl, 1 mm EDTA, 10 mm pyrophosphate, 10 mm glycerophosphate, 50 mm NaF, 0.5 mm orthovanadate) containing protease and phosphatase inhibitors. Cell lysates were incubated on ice for 35 min and then centrifuged for 20 min at 12,000 × g. Protein concentrations were determined using the BCA protein assay kit (Thermo Fisher Scientific). 30 μg of sample proteins were separated by SDS-PAGE and transferred to PVDF membrane (Millipore Corporation, Billerica, MA). Membranes were blocked with 5% fat-free milk in TBST for 1 h at room temperature and incubated with primary antibodies overnight at 4 °C and appropriate HRP-secondary antibodies for 1 h at room temperature. Protein bands were visualized with chemiluminescent agents (Beyotime, Jiangsu, China). Images were gathered by the Fusion FX6 imaging system (Vilber Lourmat, Collégien, France).
Quantitative real-time PCR analysis
Total RNA was extracted from cultured cells using TRIZOL reagent (Thermo Fisher Scientific) according to the manufacturer's protocol. First strand cDNAs were synthesized using the Moloney murine leukemia virus reverse transcriptase and oligo(dT) primers. Quantitative real-time PCR analysis was carried out on the iQ5 (Bio-Rad) using the SYBR Select Master Mix (Thermo Fisher Scientific). mRNA levels were normalized to GAPDH level. The primer sequences were as follows: GAPDH, 5′-ATGGGCAGCCGTTAGGAAAG-3′ (forward) and 5′-ATCACCCGGAGGAGAAATCG-3′ (reverse); MCL-1, 5′-TCCCTTTTCCTTGGACTGGTATC-3′ (forward) and 5′-GATGACCTTATGGCTCTGAGATGG-3′ (reverse); and GADD34, 5′-CCTCTGGCAATCCCCCATAC-3′ (forward) and 5′-TCTCGCTCACCATACATGCC-3′ (reverse).
TUNEL assay
Cell apoptosis was measured by TUNEL assay (Tsingke Biotech, Beijing, China). After being fixed in 4% paraformaldehyde for 25 min at 4 °C, the cells were incubated with proteinase K (2 mg/ml) for 10 min at room temperature. TUNEL staining was performed according to the manufacturer's instructions, followed by incubating with 4′,6′-diamino-2-phenylindole (2 μg/ml) solution at 37 °C for 5 min in dark. Quantification of all cells and apoptotic cells in same fields was performed by acquiring the images in random fields and counting cells in four random fields in each well.
Statistical analysis
One-way analysis of variance with post hoc tests was used in statistical analysis of mRNA expression and apoptosis rate. All statistical tests were two-sided, and the difference was considered statistically significant if p < 0.05.
Author contributions
P. S. data curation; P. S., S. Y., H. H., H. Z., Q. K., and T. L. investigation; P. S., Q. K., and J. W. methodology; P. S. and Y. J. writing-original draft; S. Y. and J.W. validation; H. Z. project administration; Y. J. conceptualization; Y. J. resources; Y. J. formal analysis; Y. J. supervision; Y. J. funding acquisition; Y. J. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Qiulin Tang for technical assistance.
This work was supported by Grants 81672814 and 81872388 from the National Natural Science Foundation of China and Grant 2018SCUH0009 from the Fundamental Research Fund for the Central Universities. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- ER
- endoplasmic reticulum
- HCC
- hepatocellular carcinoma
- TRAIL
- tumor necrosis factor–related apoptosis-inducing ligand
- MCL-1
- myeloid cell leukemia 1
- ERK
- extracellular signal-regulated kinase
- TRAF6
- tumor necrosis factor receptor–associated factor 6
- TAB1
- transforming growth factor-β–activated kinase 1 (MAP3K7)–binding protein 1
- eIF
- eukaryotic initiation factor
- DR
- death receptor
- CHX
- cycloheximide
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- PI3K
- phosphatidylinositol 3-kinase
- MAPK
- mitogen-activated protein kinase.
References
- 1. Emadali A., Nguyên D. T., Rochon C., Tzimas G. N., Metrakos P. P., and Chevet E. (2005) Distinct endoplasmic reticulum stress responses are triggered during human liver transplantation. J. Pathol. 207, 111–118 10.1002/path.1798 [DOI] [PubMed] [Google Scholar]
- 2. Rojas M., Vasconcelos G., and Dever T. E. (2015) An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc. Natl. Acad. Sci. U.S.A. 112, E3466–E3475 10.1073/pnas.1501557112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee J. E., Morrison W., and Hollien J. (2018) Hairy and enhancer of split 1 (HES1) protects cells from endoplasmic reticulum stress-induced apoptosis through repression of GADD34. J. Biol. Chem. 293, 5947–5955 10.1074/jbc.RA118.002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grishin A. V., Azhipa O., Semenov I., and Corey S. J. (2001) Interaction between growth arrest-DNA damage protein 34 and Src kinase Lyn negatively regulates genotoxic apoptosis. Proc. Natl. Acad. Sci. U.S.A. 98, 10172–10177 10.1073/pnas.191130798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ito S., Tanaka Y., Oshino R., Okado S., Hori M., and Isobe K. I. (2016) GADD34 suppresses lipopolysaccharide-induced sepsis and tissue injury through the regulation of macrophage activation. Cell Death Dis. 7, e2219 10.1038/cddis.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ito S., Tanaka Y., Oshino R., Aiba K., Thanasegaran S., Nishio N., and Isobe K. (2015) GADD34 inhibits activation-induced apoptosis of macrophages through enhancement of autophagy. Sci. Rep. 5, 8327 10.1038/srep08327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fransson L., Sjöholm A., and Ortsäter H. (2014) Inhibition of palmitate-induced GADD34 expression augments apoptosis in mouse insulinoma cells (MIN6). Cell Biochem. Funct. 32, 445–452 [DOI] [PubMed] [Google Scholar]
- 8. Teng Y., Gao M., Wang J., Kong Q., Hua H., Luo T., and Jiang Y. (2014) Inhibition of eIF2α dephosphorylation enhances TRAIL-induced apoptosis in hepatoma cells. Cell Death Dis. 5, e1060 10.1038/cddis.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perini G. F., Ribeiro G. N., Pinto Neto J. V., Campos L. T., and Hamerschlak N. (2018) BCL-2 as therapeutic target for hematological malignancies. J. Hematol. Oncol. 11, 65 10.1186/s13045-018-0608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gomez-Bougie P., Halliez M., Moreau P., Pellat-Deceunynck C., and Amiot M. (2016) Repression of Mcl-1 and disruption of the Mcl-1/Bak interaction in myeloma cells couple ER stress to mitochondrial apoptosis. Cancer Lett. 383, 204–211 10.1016/j.canlet.2016.09.030 [DOI] [PubMed] [Google Scholar]
- 11. Merino D., Lok S. W., Visvader J. E., and Lindeman G. J. (2016) Targeting BCL-2 to enhance vulnerability to therapy in estrogen receptor-positive breast cancer. Oncogene 35, 1877–1887 10.1038/onc.2015.287 [DOI] [PubMed] [Google Scholar]
- 12. Maji S., Panda S., Samal S. K., Shriwas O., Rath R., Pellecchia M., Emdad L., Das S. K., Fisher P. B., and Dash R. (2018) Bcl-2 antiapoptotic family proteins and chemoresistance in cancer. Adv. Cancer Res. 137, 37–75 10.1016/bs.acr.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 13. Kuo M. L., Chuang S. E., Lin M. T., and Yang S. Y. (2001) The involvement of PI 3-K/Akt-dependent up-regulation of Mcl-1 in the prevention of apoptosis of Hep3B cells by interleukin-6. Oncogene 20, 677–685 10.1038/sj.onc.1204140 [DOI] [PubMed] [Google Scholar]
- 14. Isomoto H., Kobayashi S., Werneburg N. W., Bronk S. F., Guicciardi M. E., Frank D. A., and Gores G. J. (2005) Interleukin 6 upregulates myeloid cell leukemia-1 expression through a STAT3 pathway in cholangiocarcinoma cells. Hepatology 42, 1329–1338 10.1002/hep.20966 [DOI] [PubMed] [Google Scholar]
- 15. Domina A. M., Vrana J. A., Gregory M. A., Hann S. R., and Craig R. W. (2004) MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or Taxol. Oncogene 23, 5301–5315 10.1038/sj.onc.1207692 [DOI] [PubMed] [Google Scholar]
- 16. Zhong Q., Gao W., Du F., and Wang X. (2005) Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121, 1085–1095 10.1016/j.cell.2005.06.009 [DOI] [PubMed] [Google Scholar]
- 17. Ding Q., He X., Hsu J. M., Xia W., Chen C. T., Li L. Y., Lee D. F., Liu J. C., Zhong Q., Wang X., and Hung M. C. (2007) Degradation of Mcl-1 by β-TrCP mediates glycogen synthase kinase 3–induced tumor suppression and chemosensitization. Mol. Cell. Biol. 27, 4006–4017 10.1128/MCB.00620-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Timucin A. C., Basaga H., and Kutuk O. (2019) Selective targeting of antiapoptotic BCL-2 proteins in cancer. Med. Res. Rev. 39, 146–175 10.1002/med.21516 [DOI] [PubMed] [Google Scholar]
- 19. Jeong M., Lee E. W., Seong D., Seo J., Kim J. H., Grootjans S., Kim S. Y., Vandenabeele P., and Song J. (2017) USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability. Oncogene 36, 458–470 10.1038/onc.2016.215 [DOI] [PubMed] [Google Scholar]
- 20. von Karstedt S. V., Montinaro A., and Walczak H. (2017) Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer 17, 352–366 10.1038/nrc.2017.28 [DOI] [PubMed] [Google Scholar]
- 21. Van Geelen C. M., de Vries E. G., and de Jong S. (2004) Lessons from TRAIL-resistance mechanisms in colorectal cancer cells: paving the road to patient-tailored therapy. Drug Resist. Updat. 7, 345–358 10.1016/j.drup.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 22. Farook J. M., Shields J., Tawfik A., Markand S., Sen T., Smith S. B., Brann D., Dhandapani K. M., and Sen N. (2013) GADD34 induces cell death through inactivation of Akt following traumatic brain injury. Cell Death Dis. 4, e754 10.1038/cddis.2013.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kashiwada M., Shirakata Y., Inoue J. I., Nakano H., Okazaki K., Okumura K., Yamamoto T., Nagaoka H., and Takemori T. (1998) Tumor necrosis factor receptor-associated factor 6 (TRAF6) stimulates extracellular signal-regulated kinase (ERK) activity in CD40 signaling along a ras-independent pathway. J. Exp. Med. 187, 237–244 10.1084/jem.187.2.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ge B., Gram H., Di Padova F., Huang B., New L., Ulevitch R. J., Luo Y., and Han J. (2002) MAPKK-independent activation of p38α mediated by TAB1-dependent autophosphorylation of p38α. Science 295, 1291–1294 10.1126/science.1067289 [DOI] [PubMed] [Google Scholar]
- 25. Ori D., Kato H., Sanjo H., Tartey S., Mino T., Akira S., and Takeuchi O. (2013) Essential roles of K63-linked polyubiquitin-binding proteins TAB2 and TAB3 in B cell activation via MAPKs. J. Immunol. 190, 4037–4045 10.4049/jimmunol.1300173 [DOI] [PubMed] [Google Scholar]
- 26. Yuan X., Gajan A., Chu Q., Xiong H., Wu K., and Wu G. S. (2018) Developing TRAIL/TRAIL death receptor-based cancer therapies. Cancer Metastasis Rev. 37, 733–748 10.1007/s10555-018-9728-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shlyakhtina Y., Pavet V., and Gronemeyer H. (2017) Dual role of DR5 in death and survival signaling leads to TRAIL resistance in cancer cells. Cell Death Dis. 8, e3025 10.1038/cddis.2017.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsu T. S., Mo S. T., Hsu P. N., and Lai M. Z. (2018) c-FLIP is a target of the E3 ligase deltex1 in gastric cancer. Cell Death Dis. 9, 135 10.1038/s41419-017-0165-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Safa A. R. (2012) c-FLIP, a master anti-apoptotic regulator. Exp. Oncol. 34, 176–184 [PMC free article] [PubMed] [Google Scholar]
- 30. French R., Hayward O., Jones S., Yang W., and Clarkson R. (2015) Cytoplasmic levels of cFLIP determine a broad susceptibility of breast cancer stem/progenitor-like cells to TRAIL. Mol. Cancer 14, 209 10.1186/s12943-015-0478-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hollander M. C., Zhan Q., Bae I., and Fornace A. J. (1997) Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J. Biol. Chem. 272, 13731–13737 10.1074/jbc.272.21.13731 [DOI] [PubMed] [Google Scholar]
- 32. Liu L., Ito S., Nishio N., Sun Y., Chen N., Tanaka Y., and Isobe K. (2015) GADD34 facilitates cell death resulting from proteasome inhibition. Anticancer Res. 35, 5317–5324 [PubMed] [Google Scholar]
- 33. Liu L., Ito S., Nishio N., Sun Y., Tanaka Y., and Isobe K. (2016) GADD34 promotes tumor growth by inducing myeloid-derived suppressor cells. Anticancer Res. 36, 4623–4628 10.21873/anticanres.11012 [DOI] [PubMed] [Google Scholar]
- 34. Tanaka Y., Ito S., Oshino R., Chen N., Nishio N., and Isobe K. (2015) Effects of growth arrest and DNA damage-inducible protein 34 (GADD34) on inflammation-induced colon cancer in mice. Br. J. Cancer 113, 669–679 10.1038/bjc.2015.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ding Q., Huo L., Yang J. Y., Xia W., Wei Y., Liao Y., Chang C. J., Yang Y., Lai C. C., Lee D. F., Yen C. J., Chen Y. J., Hsu J. M., Kuo H. P., Lin C. Y., et al. (2008) Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res. 68, 6109–6117 10.1158/0008-5472.CAN-08-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Inoue J., Gohda J., and Akiyama T. (2007) Characteristics and biological functions of TRAF6. Adv. Exp. Med. Biol. 597, 72–79 10.1007/978-0-387-70630-6_6 [DOI] [PubMed] [Google Scholar]
- 37. Chung J. Y., Lu M., Yin Q., Lin S. C., and Wu H. (2007) Molecular basis for the unique specificity of TRAF6. Adv. Exp. Med. Biol. 597, 122–130 10.1007/978-0-387-70630-6_10 [DOI] [PubMed] [Google Scholar]
- 38. Kotschy A., Szlavik Z., Murray J., Davidson J., Maragno A. L., Le Toumelin-Braizat G., Chanrion M., Kelly G. L., Gong J. N., Moujalled D. M., Bruno A., Csekei M., Paczal A., Szabo Z. B., Sipos S., et al. (2016) The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482 10.1038/nature19830 [DOI] [PubMed] [Google Scholar]
- 39. Kim S. Y., Park S. E., Shim S. M., Park S., Kim K. K., Jeong S. Y., Choi E. K., Hwang J. J., Jin D. H., Chung C. D., and Kim I. (2015) Bay 61–3606 sensitizes TRAIL-induced apoptosis by downregulating Mcl-1 in breast cancer cells. PLoS One 10, e0146073 10.1371/journal.pone.0146073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lemke J., von Karstedt S., Abd El Hay M., Conti A., Arce F., Montinaro A., Papenfuss K., El-Bahrawy M. A., and Walczak H. (2014) Selective CDK9 inhibition overcomes TRAIL resistance by concomitant suppression of cFlip and Mcl-1. Cell Death Differ. 21, 491–502 10.1038/cdd.2013.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murphy Á. C., Weyhenmeyer B., Noonan J., Kilbride S. M., Schimansky S., Loh K. P., Kögel D., Letai A. G., Prehn J. H., and Murphy B. M. (2014) Modulation of Mcl-1 sensitizes glioblastoma to TRAIL-induced apoptosis. Apoptosis 19, 629–642 10.1007/s10495-013-0935-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schewe D. M., and Aguirre-Ghiso J. A. (2009) Inhibition of eIF2α dephosphorylation maximizes bortezomib efficiency and eliminates quiescent multiple myeloma cells surviving proteasome inhibitor therapy. Cancer Res. 69, 1545–1552 10.1158/0008-5472.CAN-08-3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ebinger S., Özdemir E. Z., Ziegenhain C., Tiedt S., Castro Alves C., Grunert M., Dworzak M., Lutz C., Turati V. A., Enver T., Horny H. P., Sotlar K., Parekh S., Spiekermann K., Hiddemann W., et al. (2016) Characterization of rare, dormant, and therapy-resistant cells in acute lymphoblastic leukemia. Cancer Cell 30, 849–862 10.1016/j.ccell.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.