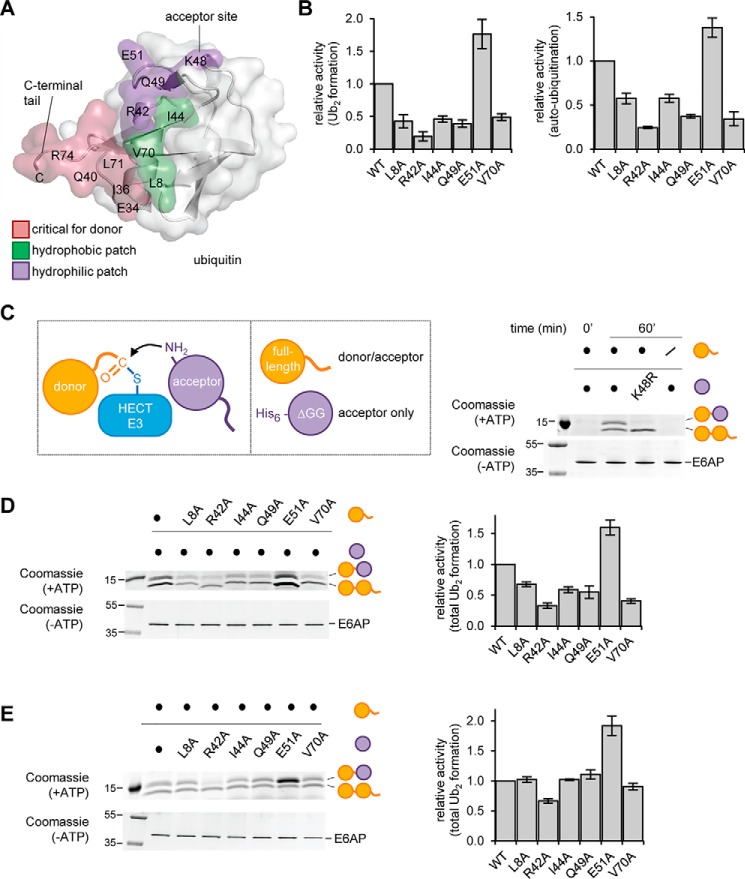
Figure 9.
Distinct surface regions are required by the acceptor and donor ubiquitin during E6AP-catalyzed ubiquitin linkage formation. A, structure of ubiquitin (PDB code 1UBQ (94)) is shown in ribbon and surface representation. The area of the donor ubiquitin, including the C-terminal tail, that contacts the HECT C-lobe in the canonical mode is colored rose; hydrophobic residues important for E6AP activity are in green (49); and residues in proximity of Lys-48, as studied in B–D, are in purple. B, quantification of the isopeptide bond formation activity of the E6AP HECT domain toward ubiquitin variants, based on three independent experiments (for experimental data, see Fig. S5). For free chain formation (left) and auto-ubiquitination (right), the amount of reaction product (Ub2 and E6APUb, respectively) at 60 min was normalized to that of input E6AP. C, schematic of HECT ligase-mediated linkage formation between an enzyme-linked donor and an acceptor ubiquitin (left), and the two ubiquitin substrates employed in the ΔGG assay: WT ubiquitin and His6-tagged, truncated ubiquitin (residues 1–74; UbΔGG) (middle); validation of the assay using the E6AP HECT domain and equimolar mixtures of the indicated ubiquitin substrates. The Ub2 reaction products were analyzed by reducing SDS-PAGE and Coomassie staining. The input amount of E6AP in the absence of ATP serves as a control (right). D and E, ΔGG assay with ubiquitin variants, performed and analyzed as in C. The amount of both Ub2 products combined were quantified and normalized to the amount of input E6AP, and the means and standard deviations from three independent experiments were plotted (right).