Figure 8.
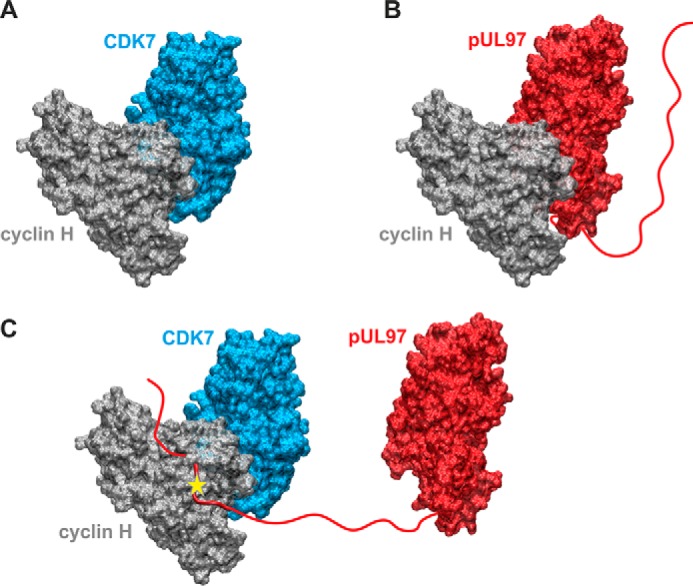
Structural model of the putative cyclin H-bridged interaction between pUL97 and CDK7 as presented in Fig. 7. A, model of the binary CDK7–cyclin H complex. B, model of the binary pUL97–cyclin H complex. Both models were generated based on the homologous CDK9–cyclin T1 complex crystal structure using the strategy described before (14) and show the canonical interaction via the large globular domain interfaces. The globular domain interfaces shown in A and B are overlapping, and thus only one partner can interact with cyclin H via this interface. C, model of a putative ternary CDK7–cyclin H-pUL97 complex, in which the long unstructured N-terminal region of pUL97 is supposed to interact with cyclin H via an alternative binding motif or interface. A yellow star denotes a yet undefined modification of cyclin H induced by the HCMV-specific infectious environment that is required for pUL97 binding. Note that this figure shows the minimal composition of such a ternary complex containing one copy of each component. However, it is possible that higher-order complexes may contain additional copies of some components (e.g. two cyclin H molecules bound to pUL97, one via its N-terminal binding motif, a second via its globular domain interface, see B).
