Abstract
T-cell receptors (TCRs) recognize pathogens to ignite immune responses, making them attractive scaffolds for development as immunotherapeutics. However, manipulation of TCRs has been impeded by difficulties in their engineering and expression. Wagner and colleagues now establish new platforms to generate high-affinity TCR variants that potently activate T cells, and they also create soluble TCR fusion proteins that specifically recognize cognate peptides. This work provides specific tools to combat cytomegalovirus (CMV) infection and helps illuminate a general path to actuation of engineered TCR-based therapeutics.
Introduction
Antibodies have become one of the most powerful weapons in immunotherapy over the past 25 years due to their pinpoint target specificity, modular construction, favorable pharmacokinetic properties, and multilayered mechanisms of action (1). Antibodies act through target neutralization and immune effector cell recruitment, but they can also be engineered for other purposes, such as in the design of diagnostics, for drug delivery, as bispecific ligands, and for the modulation of host biology (2). T-cell receptors (TCRs)3 share many biochemical and structural features with antibodies, and they also share the functional capacity to specifically recognize antigens, albeit through a different mode of recognition. Whereas antibodies directly engage antigens, TCRs interact with cognate antigenic peptides presented by major histocompatibility complex (MHC) molecules on the surface of antigen-presenting cells (APCs). Because peptides associated with MHC class I molecules are commonly derived from intracellular proteins, TCRs can access both cell surface and intracellular antigens, expanding their potential as targeted drugs (3). However, therapeutic translation of TCRs has been stifled by complications with engineering and expression. Wagner and colleagues (4) now provide new methods to overcome these challenges and employ them to generate a soluble TCR molecule with nanomolar affinity for an antigen associated with pathogenic human cytomegalovirus (CMV). These technologies harbor important implications for advancing the development of TCR-based therapies.
Despite the vast utility TCRs could provide in diversifying target recognition, several obstacles have prevented TCRs from reaching the same level of therapeutic prominence as antibodies. First, TCR binding affinities are much weaker than those of antibodies; thus, engineering is required to boost their affinities to a therapeutically useful range. Like antibodies, TCR binding sites comprise six hypervariable complementarity-determining regions (CDRs), three each on the α- and β-chains (5). Prior engineering efforts have employed phage or yeast display to evolve TCRs with higher affinity, but these systems do not include the proper folding or glycosylation machinery needed to present complex mammalian proteins. TCRs have previously been studied using mammalian display systems, which relied on lentiviral transduction (6, 7), but this process is very slow, and the number of variants that can be examined is low. Finally, since the α- and β-chains of TCRs are embedded in the membrane, their transmembrane sequences must be removed for any exogenous application, and no general solutions to this problem exist.
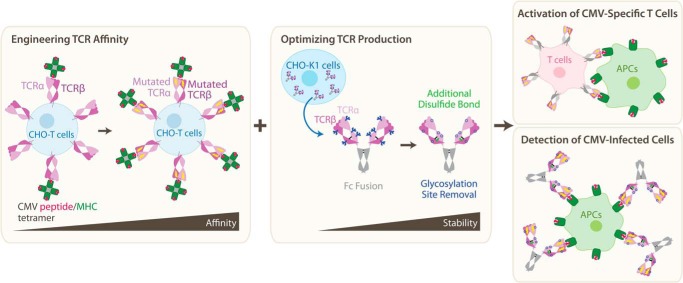
To address these challenges, Wagner and colleagues (4) establish a platform for presenting the TCR α- and β-chains on the surface of mammalian cells, introducing an inter-chain disulfide bond to enhance stability. This system was used to evolve a TCR to bind a CMV-associated peptide–MHC complex with 60-fold higher affinity (Fig. 1). Specifically, the authors used a sequence from the CMV matrix protein pp65 (NLVPMVATV) known to be loaded in the MHC HLA-A*02 allele (8); the authors denote this complex as NLV/A2. Hundreds of natural TCRs recognize NLV/A2, allowing the identification of conserved recognition motifs. One clone (RA14) contains the two most conserved sequences in its CDR3 domains (9) and was thus used as the basis for engineering CMV-targeted TCRs. The authors designed two separate libraries of the α- and β-chains with mutagenized residues in the CDR3 domains. Iterative rounds of fluorescence-activated cell sorting (FACS) against NLV/A2 yielded both α- and β-chain variants with improved affinity. The mutated α- and β-chains were combined pairwise to identify the optimal final variant, denoted α2.β8. Recent work revealed TCR affinity to be less predictive of T-cell function than expected (10). However, transfection of the α2.β8 TCR variant into human Jurkat T cells led to specific activation by the NLV peptide presented by APCs that express the A2 MHC, and this activation was significantly more potent than that induced by the WT RA14 (Fig. 1).
Figure 1.
To overcome limitations in TCR engineering, the authors established new mammalian cell-based platforms for enhancing receptor affinity (left) and soluble production (center). These platforms generated TCRs that potently and specifically recognized APC-presented CMV antigens both in transduced T cells (right top) and as soluble TCR–antibody fusion proteins (right bottom).
Wagner and colleagues (4) next tackled the issue of protein expression. Although prior work has described TCR-antibody chimeras, these constructs have not resolved issues of low expression levels and poor assembly. Here, the authors report a simplified design, in which the antigen-binding domain of the TCR is fused to constant domains of an antibody. Furthermore, an additional non-native disulfide bond was introduced, and predicted N-linked glycosylation sites near domain interfaces were removed, as these could sterically inhibit assembly. These modifications improved protein yield and resulted in a higher melting temperature, comparable to that of antibodies. The authors then incubated APCs expressing NLV/A2 with TCR–antibody fusion proteins and found that whereas no staining was observed for WT RA14, the α2.β8 mutant exhibited robust staining (Fig. 1), supporting the potential use of engineered TCRs in detection of CMV-infected cells.
The bulk of previous TCR engineering has focused on modulating cancer antigen affinity. This work is one of few examples in which a human TCR that recognizes an infectious disease-related peptide, aside from HIV, was successfully engineered to have increased affinity and induce enhanced activation. Furthermore, many prior examples of affinity-maturing TCRs resulted in loss of specificity, allowing activation by noncognate peptides. Here, the affinity-matured TCR maintained its specificity, suggesting that transduction of engineered TCRs could prove useful in cellular therapies against CMV, particularly in immunocompromised individuals for whom the virus can prove fatal. The soluble TCR–antibody fusion protein may also have therapeutic potential on its own, by binding to CMV-infected cells and thereby tagging those cells for immune clearance, and could provide a simpler diagnostic method to assess infection status.
Perhaps most notably, the authors' unique TCR display format and the use of CHO-T cells as their expression and selection platform allow for rapid transfection of the TCR and stable presentation with adequate time to complete multiple rounds of selection on the TCR library. This represents a powerful technique for comparatively rapid TCR engineering and enables analysis of constructs that are more relevant to human physiology, while eliminating potential downstream manufacturing concerns that could result from the differential post-translational modifications in nonmammalian expression systems. Future use of these novel platforms will assist in probing the interplay between peptide–MHC/TCR interaction strength and T-cell function.
This work was supported by Department of Defense Grant RT170165 and a V Foundation Scholars award (to J. B. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TCR
- T-cell receptor
- CDR
- complementarity-determining region
- CMV
- cytomegalovirus
- MHC
- major histocompatibility complex
- APC
- antigen presenting cell
- FACS
- fluorescence-activated cell sorting.
References
- 1. Grilo A. L., and Mantalaris A. (2019) The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 37, 9–16 10.1016/j.tibtech.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 2. Weiner G. J. (2015) Building better monoclonal antibody-based therapeutics. Nat. Rev. Cancer 15, 361–370 10.1038/nrc3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L., and Flies D. B. (2013) Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 10.1038/nri3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wagner E. K., Qerqez A. N., Stevens C. A., Nguyen A. W., Delidakis G., and Maynard J. A. (2019) Human cytomegalovirus-specific T-cell receptor engineered for high affinity and soluble expression using mammalian cell display. J. Biol. Chem. 294, 5790–5804 10.1074/jbc.RA118.007187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rossjohn J., Gras S., Miles J. J., Turner S. J., Godfrey D. I., and McCluskey J. (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 10.1146/annurev-immunol-032414-112334 [DOI] [PubMed] [Google Scholar]
- 6. Chervin A. S., Aggen D. H., Raseman J. M., and Kranz D. M. (2008) Engineering higher affinity T cell receptors using a T cell display system. J. Immunol. Methods 339, 175–184 10.1016/j.jim.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Malecek K., Zhong S., McGary K., Yu C., Huang K., Johnson L. A., Rosenberg S. A., and Krogsgaard M. (2013) Engineering improved T cell receptors using an alanine-scan guided T cell display selection system. J. Immunol. Methods 392, 1–11 10.1016/j.jim.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wills M. R., Carmichael A. J., Mynard K., Jin X., Weekes M. P., Plachter B., and Sissons J. G. (1996) The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70, 7569–7579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trautmann L., Rimbert M., Echasserieau K., Saulquin X., Neveu B., Dechanet J., Cerundolo V., and Bonneville M. (2005) Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 175, 6123–6132 10.4049/jimmunol.175.9.6123 [DOI] [PubMed] [Google Scholar]
- 10. Sibener L. V., Fernandes R. A., Kolawole E. M., Carbone C. B., Liu F., McAffee D., Birnbaum M. E., Yang X., Su L. F., Yu W., Dong S., Gee M. H., Jude K. M., Davis M. M., Groves J. T., Goddard W. A. 3rd, Heath J. R., Evavold B. D., Vale R. D., and Garcia K. C. (2018) Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell 174, 672–687.e27 10.1016/j.cell.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]