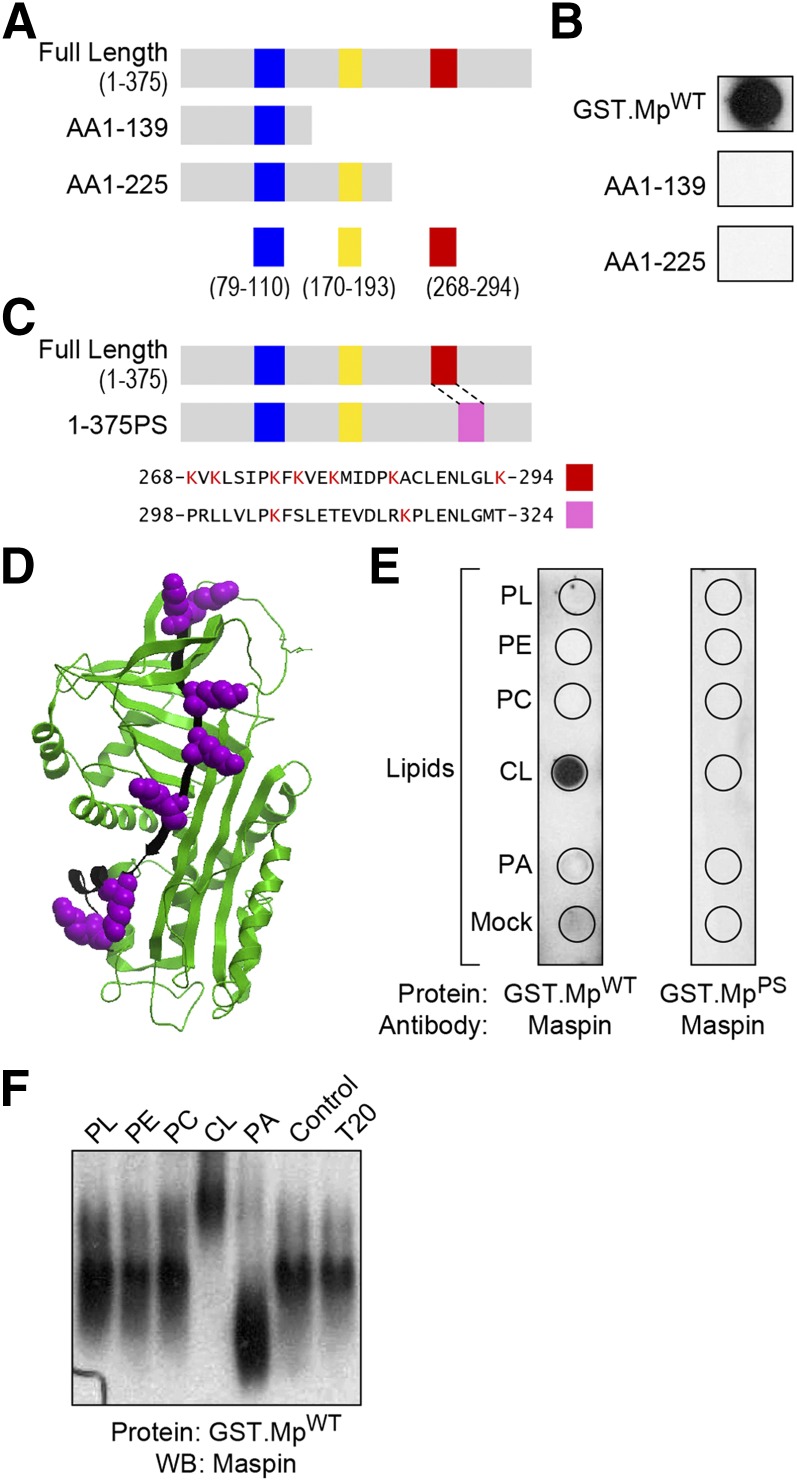
Figure 2.
Maspin binds CL through a CLBD. A) A schematic representing 6 different putative CLBDs (highlighted with blue, yellow, and red bars) in full-length maspin and truncated mutants containing protein sequences rich in hydrophobic amino acids and Lys residues. B) Comparison of the binding of different constructs of maspin to CL using a protein-lipid overlay assay. Only MpWT bound to CL. C) A schematic depicting a maspin construct in which the 7 Lys residues in the CLBD (region highlighted in red) were swapped with the corresponding sequence from the serpin family member, PAI-1 (pink box). A sequence alignment of maspin (AAA18957.1) and PAI-1 (AAA60009.1) demonstrates the homology of these 2 domains in which 7 Lys residues are present in this region of maspin (red box, aa 268–294) and 2 Lys residues in PAI-1 (pink box, aa 298–324). D) Three-dimensional structure of maspin (Protein Data Bank ID: 1XU8) highlighting the CLBD (depicted in black) and Lys residues (depicted in purple) using Internal Coordinate Mechanics (ICM) Pro v.3.48 software. E) A comparison of binding of GST.MpWT and GST.MpPS with various lipids [phospholipids (PL), phosphatidylethanolamine (PE), PC, CL, phosphatidic acid (PA), BSA (mock)] demonstrates the importance of the CLBD in maspin for binding CL. F) PAGE-Western blots demonstrate that CL causes a gel shift of wild-type maspin (GST.MpWT). Representative figures are shown of experiments completed a minimum of 6 times.