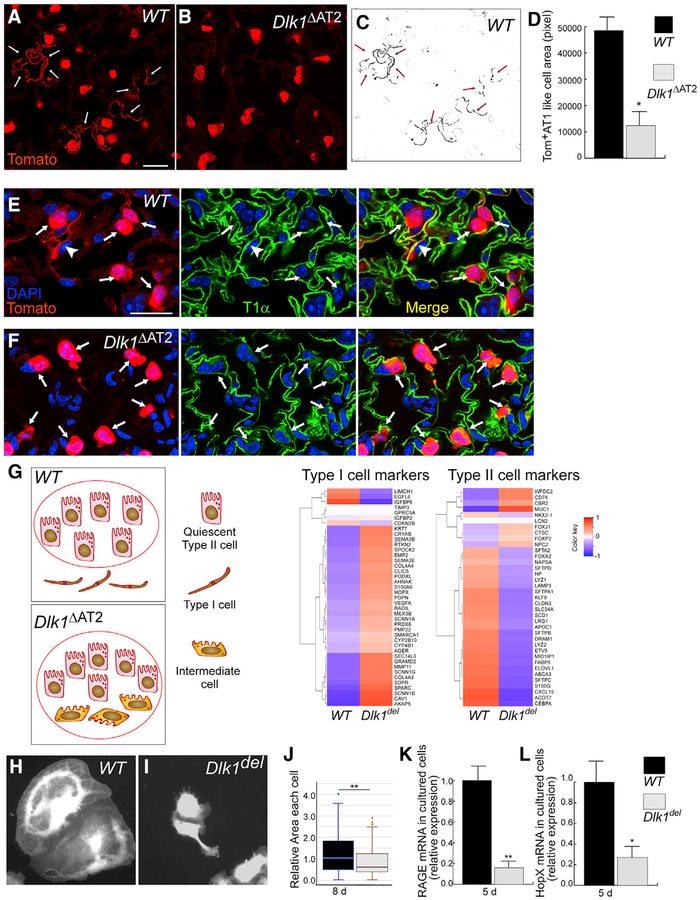
Figure 5. Dlk1del AT2 Show Defective Transition to AT1.
(A and B) Lung sections from 10-day post-PA WT (A) and Dlk1ΔAT2 (B) mice. Tomato lineage tracing lungs were stained for Tomato to mark for AT2 as well as AT1 resulting from transition of AT2. More Tomato+ AT1 were observed in WT than Dlk1ΔAT2 lungs, as marked by white arrows.
(C and D) The area of Tomato+ AT1 (arrows in C) was quantified (D) as pixel number and normalized to the number of Tomato+ AT2 of the same section. (C) represents the Tomato+ lineage-traced AT1-like cells in (A). Scale bar, 20 μm. Data are presented as mean ± SE; *p < 0.05; n = 3 mice per group. For each mouse, more than 10 microscope areas were randomly selected from sections for quantification.
(E and F) Mutant AT2-like cells expressed low levels of T1α 10 days after PA injury. Sections of AT2-lineage-traced WT (E) and Dlk1ΔAT2 (F) lungs were prepared 10 days after PA injury and processed for antibody staining of the AT1 marker T1α (green). Arrows show Tomato+ cuboidal AT2-like cells, and the arrowheads in (E) show Tomato lineage-labeled AT1. Numerous AT2-like cells in the mutant expressed a low level of T1α (arrows in F).
(G) Tomato+ AT2 and AT2-like cells were isolated from WT and Dlk1ΔAT2 lungs 9 days after PA infection and processed for RNA-seq analysis. The cartoon shows the possible types of Tomato+ lineage-traced cells in WT and Dlk1ΔAT2 lungs. Tomato+ cells were sorted by FACS following the standard AT2 isolation procedure and then processed for RNA-seq. A red circle indicates the cells included in the RNA-seq analysis. In the WT, only quiescent AT2 were included; the lineage-traced AT1 were not included in the cell preparation. In the mutant, quiescent AT2 as well as the cuboidal AT2-like intermediate cells were included. The expression levels of ~40 AT1 and AT2 markers each were compared between WT and Dlk1del cells. A modified Z score, calculated based on the log2 scale mean values, was plotted in heatmaps (n = 3 mice in each group). Red to blue coloring represents high to low expression of genes (NCBI GEO: GSE 124259).
(H–J) Tomato+ AT2 were isolated from WT (H) and Dlk1ΔAT2 (I) lungs and cultured for 8 days. WT cells (H) spread to a larger area compared with Dlk1del cells (I). The area of each cell was measured by ImageJ and is presented as a box whisker plot (J). More than 100 cells were measured for each sample group in 2 independent experiments.
(K and L) AT2 isolated from WT and Dlk1ΔAT2 lungs were cultured for 5 days to allow AT2 to AT1 transition and processed for qRT-PCR to examine expression levels of the AT1 markers RAGE (K) and HopX (L). n = 4.
Data are presented as mean ± SE; *p < 0.05, **p < 0.01. See also Figure S5.