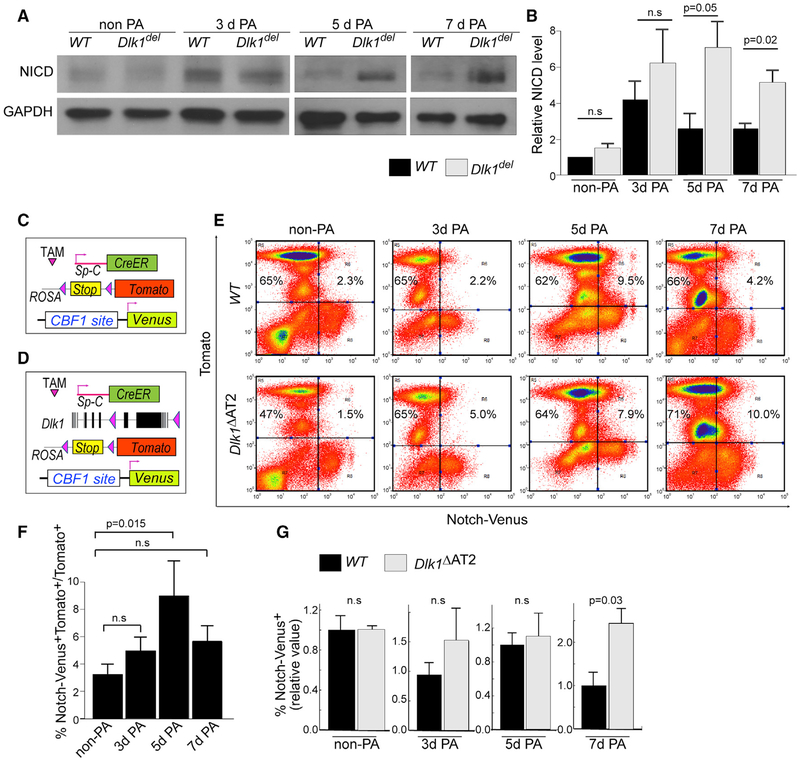
Figure 6. Sustained Notch Activation in Dlk1del AT2.
(A) Tomato+ AT2 were sorted from uninjured and 3-, 5-, and 7-day post-injury lineage-traced mice for western blot analysis of cleaved NICD. GAPDH was used as a loading control. The images of 5- and 7-day PA-injured mice are from the same gel.
(B) Intensities of western bands were quantified using ImageJ and normalized to GAPDH levels. Relative NICD levels were calculated by comparison with normalized NICD of non-PA WT samples run at the same time in each experiment. Bar graphs are presented as mean ± SE; n ≥ 3 for each genotype at each time point.
(C and D) Schematic of WT (C) or Dlk1ΔAT2 (D) lines combined with Tomato lineage tracing and the Notch activity reporter allele Notch-Venus.
(E) FACS analysis of Notch-Venus+ cells among Tomato+ AT2 of WT and mutant mice uninjured as well as 3-, 5-, and 7-day post-PA injury.
(F) Percentages of Venus+ cells among Tomato+ AT2 in WT mice non-PA-injured and 3, 5, and 7 days after PA injury were plotted.
(G) Relative ratios of Venus+ AT2 of the mutant compared with the WT non-PA-injured and 3, 5, and 7 days after PA were plotted.
Data were first calculated as percent Venus+Tomato+ cells versus Tomato+ cells, which were then compared with percent in control mice of the same PA challenge experiment. Mean ± SE; n ≥ 3. The p values are indicated. n.s., not significant. See also Figure S6.