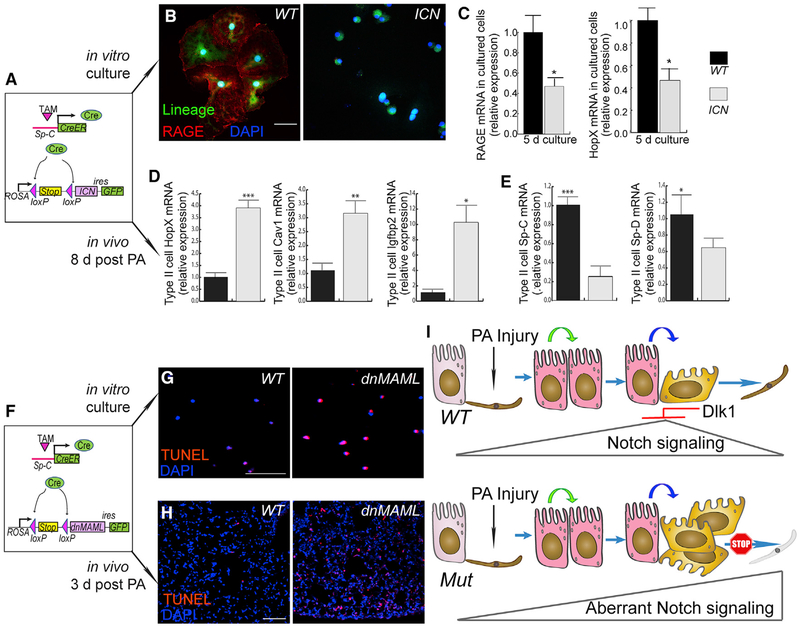
Figure 7. Dysregulated Notch Activation Disrupts AT2-to-AT1 Transition.
(A) Schematic of the mouse line with constitutively active Notch in AT2. The Notch 1 intracellular domain (ICN) was used to achieve constitutive Notch and is expressed through AT2-specific Cre recombination and the ROSA/loxP-stop-loxP locus; GFP was expressed through an IRES site as a lineage-tracing marker.
(B) AT2 from WT or SPC-ICN mice were cultured for 8 days and then stained for the AT1 marker RAGE. Green indicates the SpC-CreER-dependent AT2 lineage marker. Most lineage-labeled cells in the WT show RAGE staining (red, left), whereas almost all ICN-expressing cells (GFP+, right) were absent of RAGE.
(C) AT2 from SPC-ICN mice were cultured for 5 days and processed for qRT-PCR analysis for the appearance of the AT1 markers RAGE and HopX. Data are presented as mean ± SE; n = 3; *p < 0.05.
(D and E) Lineage-labeled AT2 were isolated by flow-sorting from WT or SPC-ICN mouse lungs 8 days after PA injury and processed for qRT-PCR analysis for expression of the AT1 markers HopX, Cav1, and Igfbp2 (D) as well as the AT2 markers Sp-C and Sp-D (E). Mean ± SE; n = 4–5; *p < 0.05, **p < 0.01, ***p < 0.001.
(F) Schematic of mouse lines with disrupted Notch signaling in AT2. Dominant-negative MAML (dnMAML) was expressed through SpC-CreER and the ROSA/loxP-stop-loxP locus, and GFP was expressed through an IRES site as a lineage-tracing marker.
(G) AT2 from WT (left) or SPC-dnMAML (right) mice were cultured for 24 h and processed for TUNEL staining.
(H) Lung sections from WT (left) or SPC-dnMAML (right) mice were prepared 3 days after PA infection and processed for TUNEL staining. Scale bar, 50 μm.
(I) The model. Upon PA injury, Notch signaling is activated in AT2. Notch signaling is later inhibited through Dlk1 before transitioning to AT1. In the absence of Dlk1, Notch signaling persists, and cells are arrested in an intermediate Sp-Clow/T1αlow stage.
See also Figure S7.