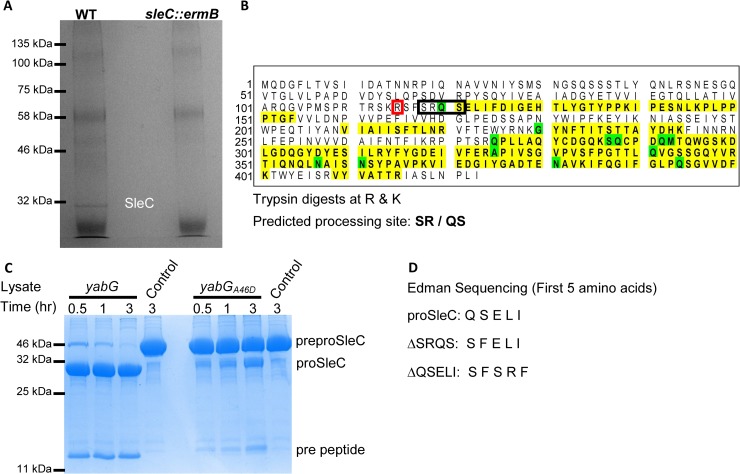
Fig 6. Identifying and characterizing the YabG processing site in preproSleC.
(A) Spores derived from C. difficile R20291 (WT) and C. difficile RS10 (sleC::ermB) were disrupted by bead-beating and then proSleC was immunoprecipitated using SleC-specific antisera. Samples were separated by SDS-PAGE gel and stained with Coomassie blue. The band corresponding to proSleC was excised from the gel and analyzed by peptide mass finger printing. (B) The sequence of preproSleC is listed. Yellow highlighted regions correspond to fragments that were detected by mass spectrometry and amino acids highlighted in green indicate amino acids that are modified in the MS analysis (i.e., oxidized, deaminated, acetylated or ammonia loss). (C) Recombinantly expressed and purified preproSleC was incubated for the indicated times in E. coli lysate that expressed either wildtype yabG or yabGA46D (Table 2). A control sample of recombinant protein incubated in buffer was included. Samples were boiled in SDS-sample buffer and separated by SDS-PAGE before staining with Coomassie Blue. (D) Recombinantly expressed and purified preproSleC, preproSleCΔSRQS or preproSleCΔQSELI were incubated with wildtype YabG as in (C). The first five amino acids at the N-terminus were determined by Edman sequencing. R115 is highlighted in red.