Figure 5.
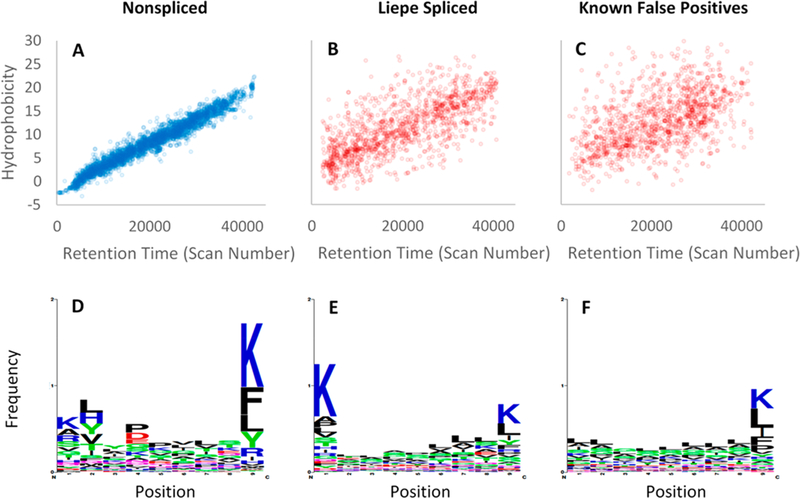
Comparison of peptide identifications previously reported by Liepe et al. for HLA-I-Ip of primary fibroblasts. (A) Nonspliced identifications show tight correlations between SSRCalc predicted HI and observed RT. (B) LM_PSPs show a weak correlation between HI and RT, indicating an underestimated FDR. (C) Known false positive PSMs are displayed to visualize the poor correlation between HI and RT for incorrect identifications. The false positives PSMs are the top scoring decoys from a MetaMorpheus search, where the number of decoys plotted is equal to the number of LM_PSPs. (D) The nonspliced sequences have common residues at the N2 (#2) and C1 (#9) anchor positions where the antigens bind to HLA-I. The LM_PSPs (E) and decoy identifications (F) display a weaker C1 specificity and an absence of N2 specificity.
