Figure 1. Overview of the read cloud shotgun sequencing and assembly approach.
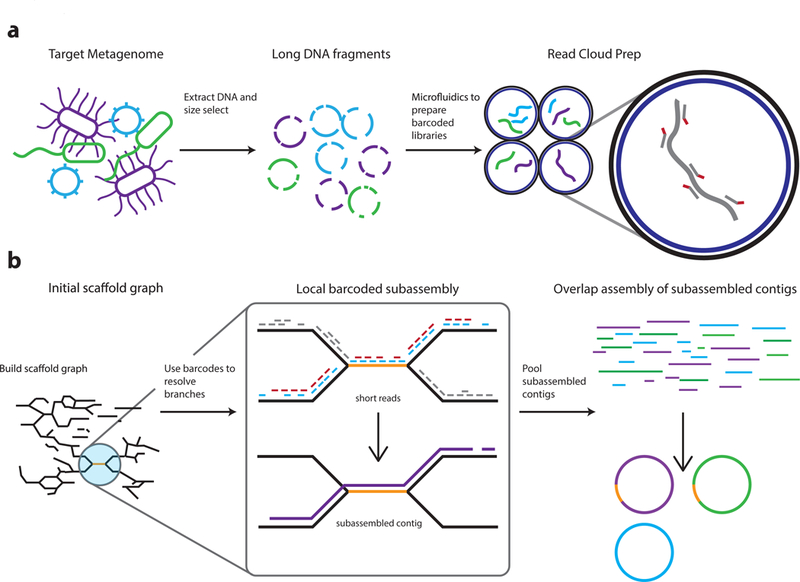
a) DNA is first extracted from microbiome samples and is size selected to enrich for long DNA fragments. The long fragments are then diluted and undergo sparse partitioning across more than a million droplet partitions (using, for example, the 10X Genomics Chromium library preparation platform). Degenerate amplification of these long fragments is then performed within these partitions to obtain barcoded traditional libraries -- each with a barcode unique to its partition. These libraries are then pooled and sequenced with an Illumina instrument.
b) The Athena assembler uses read clouds to yield more complete drafts in which genomic repeats are also accurately placed. An example repeat that is resolved and placed by Athena is shown in orange. 1) Read clouds are first assembled with standard short-read techniques to obtain seed contigs, input reads are mapped back to these seed contigs, and read pairs that span two seed contigs are used to build a scaffold graph containing unresolvable branches. 2) At each edge, Athena proposes a much simpler subassembly problem on a pooled subset of barcoded reads informed by the scaffold graph mappings. Example short reads with red and blue barcodes are passed to a short-read assembler to perform subassembly, which yields a longer subassembled contig that disambiguates branches in the scaffold graph. 3) The resulting subassembled contigs, together with the initial seed contigs, are then passed as reads to the long read De Bruijn graph based assembler Flye for final assembly. The resulting draft assembly metagenome produces more complete and more contiguous drafts in which repeats are also assembled and correctly placed.
