Figure 3. Combined genome draft results of read cloud, SLR, and short read approaches applied to healthy human stool samples.
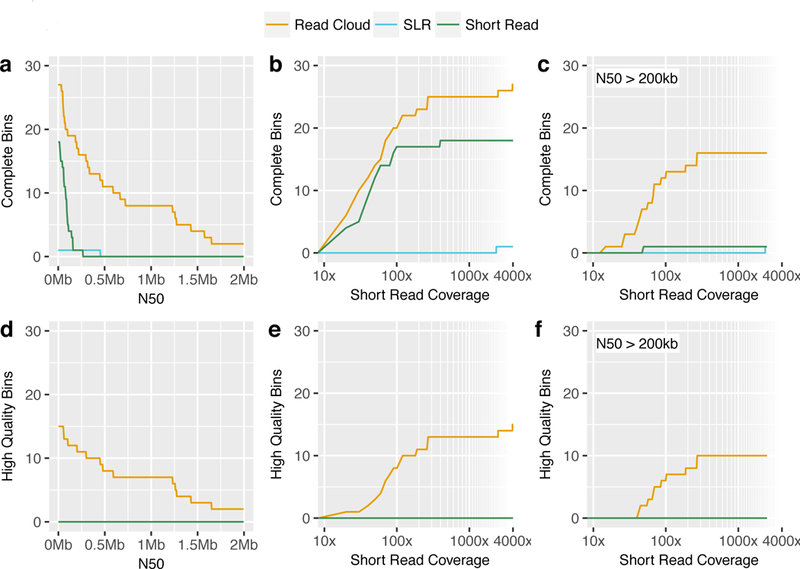
Under various performance metrics, read clouds (gold) consistently display superior performance in their ability to produce many complete and high-quality genome drafts as compared to either SLRs (blue) or short reads (green) approaches. Performance was also superior even in low short read coverage regimes (defined as <50x coverage). Counts include all complete/high-quality genome bins for all taxa in each approach.
a) Number of complete genome bins (>90% completeness, <5% contamination) with a minimum N50.
b) Number of complete genome bins with a minimum short read coverage depth. Genome bins with lower short read coverage correspond to less abundant organisms.
c) Number of complete genome bins with an N50 of >200kb and a minimum short read coverage depth.
d) Number of high-quality genome bins (complete and with at least 18 tRNAs, as well as at least one instance each of the 5S, 16S, and 23S rRNA genes) with a minimum N50.
e) Number of high-quality genome bins with a minimum short read coverage depth.
f) Number of high-quality genome bins with an N50 of >200kb and a minimum short read coverage depth.
