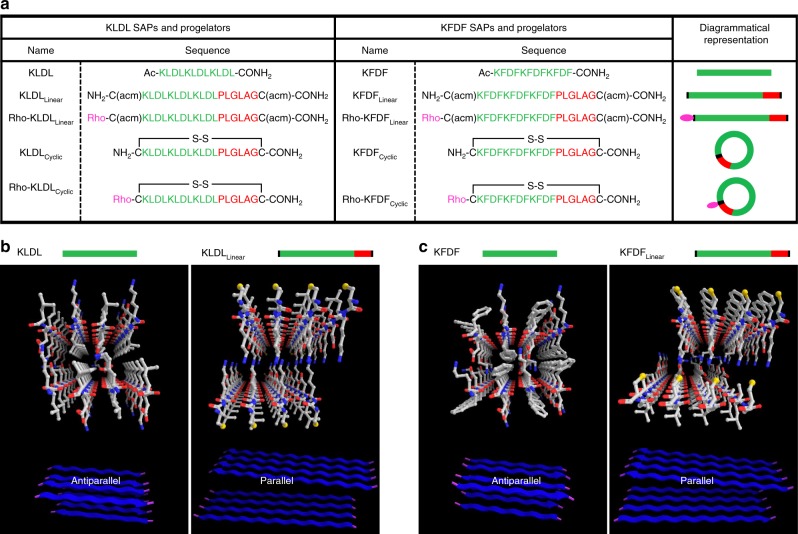
Fig. 2.
Sequences and design of self-assembling peptides (SAPs) and cyclic, enzyme-responsive progelators. a Table of SAPs and progelators based on the (KLDL)3 and (KFDF)3 amino acid gelling sequence with corresponding diagrammatical representations (green is progelator sequence, red is recognition sequence, pink is fluorophore). Sequences are shown for the base, unmodified SAPs (KLDL and KFDF) and modified SAPs (KLDLLinear and KFDFLinear) primed for cyclization but lacking fluorescent labels, together with the corresponding cyclized versions (progelators, KLDLCyclic and KFDFCyclic). In addition, rhodamine-labeled modified SAPs (Rho-KLDLLinear and Rho-KFDFLinear) are shown, together with their corresponding labeled cyclized progelators (Rho-KLDLCyclic and Rho-KFDFCyclic). The ‘acm' denotes the acetamidomethyl protecting group on Cysteine residues. ‘Rho' designates the 5(6)-carboxytetramethyl rhodamine label. b, c Modeling predicts β-sheet re-orientation, rather than self-assembly disruption, following significant SAP functionalization. b Predicted structures and ribbon diagrams (bottom) of KLDL and KLDLLinear SAPs from FibPredictor simulations revealing antiparallel and parallel β-sheet orientations, respectively. c Predicted structures and ribbon diagrams (bottom) of KFDF and KFDFLinear SAPs from FibPredictor simulations revealing antiparallel and parallel β-sheet orientations, respectively (see Supplementary Tables 1-2). Ball and stick models of interacting peptides are displayed with carbon, oxygen, and nitrogen atoms in white, red, and blue, respectively