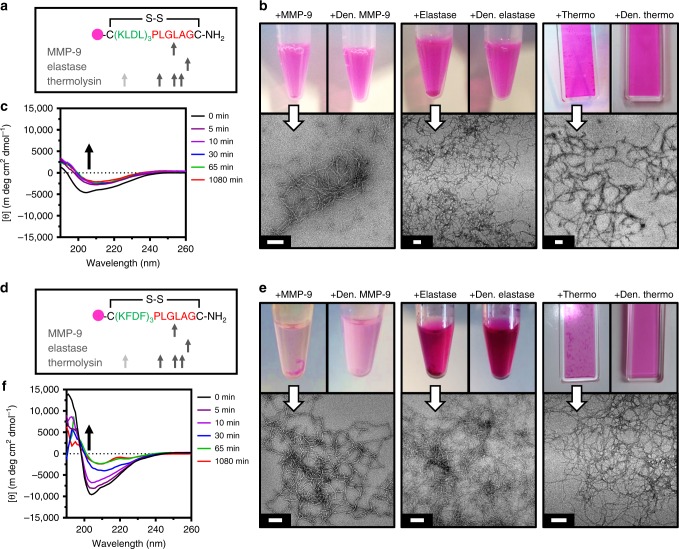
Fig. 5.
Enzyme responsiveness of Rho-KLDLCyclic and Rho-KFDFCyclic progelators. a–c Analysis of Rho-KLDLCyclic responsiveness to matrix metalloproteinase-9 (MMP-9), elastase, and thermolysin. a Peptide sequence and theoretical enzymatic cuts sites. Gray arrow (not observed). b Photographs (top) of progelator incubated with active enzymes (left insets) shows material aggregation and settling from solution, and progelator incubated with denatured enzymes (right insets) show fully dispersed peptide solutions. Corresponding transmission electron microscopy (TEM) images (bottom) of active enzyme cleavage products show fiber formation. Scale 100 nm. c Circular dichroism (CD) spectra of cleavage kinetics with thermolysin. Disappearance of signal at 204 nm (black arrow), corresponds to ring-opening. d–f Analysis of Rho-KFDFCyclic to responsiveness MMP-9, elastase, and thermolysin. d Peptide sequence and theoretical enzymatic cuts sites. Gray arrow (not observed). e Photographs (top) of progelator incubated with active enzymes (left insets) shows material aggregation and settling from solution, and progelator incubated with denatured enzymes (right insets) show fully dispersed peptide solutions. Corresponding TEM images (bottom) of active enzyme cleavage products show fiber formation. Scale 100 nm. f CD spectra of cleavage kinetics with thermolysin. Disappearance of signal at 204 nm (black arrow) corresponds to ring opening. CD of 500 µM progelator in 10 mM Tris buffer, pH 7.4. Enzyme cleavages performed at 500 µM progelator with 1:1000, 1:250, and 1:4500 enzyme/substrate molar ratio in 1× cleavage buffers (see Methods). Samples diluted to 100 µM for TEM