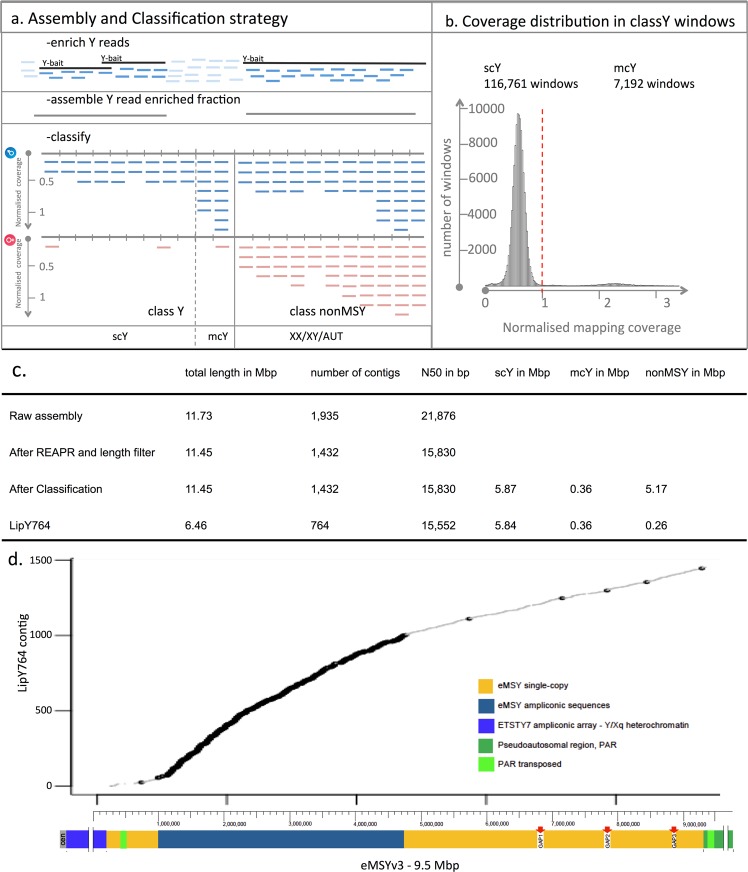
Figure 1.
LipY764 Assembly. (a) Assembly step: whole genome NGS reads from males (blue) are mapped to Y-specific bait sequences (black). Mapped reads (dark blue) are then extracted and assembled (grey). Classification step: the assembly is shown in grey with hatchmarks representing 50 bp windows. Male (blue) and female (red) reads are mapped to the assembly and mapping coverages normalised to autosomal coverages per window were estimated. The probability of Y- or nonMSY-specificity per window is obtained by comparing normalised coverages in males and females. class nonMSY: XX/XY/AUT ≈ 1 in males and ≈ 1 in females; class Y: scY ≤ 1 in males and ≈ 0 in females, mcY > 1 in males and ≈ 0 in females. (b) Frequency distribution of normalised mapping coverage in classY windows. The cut-off scY to seperate mcY is set to 1 (red dashed line). (c) Resulting statistics for the assembly and classification approach. (d) Position of LipY764 contigs on eMSYv346 (data in Supplementary Table S5). Contigs having a single unique position on eMSYv3 are shown in grey, contigs with multiple hits in black/bold.