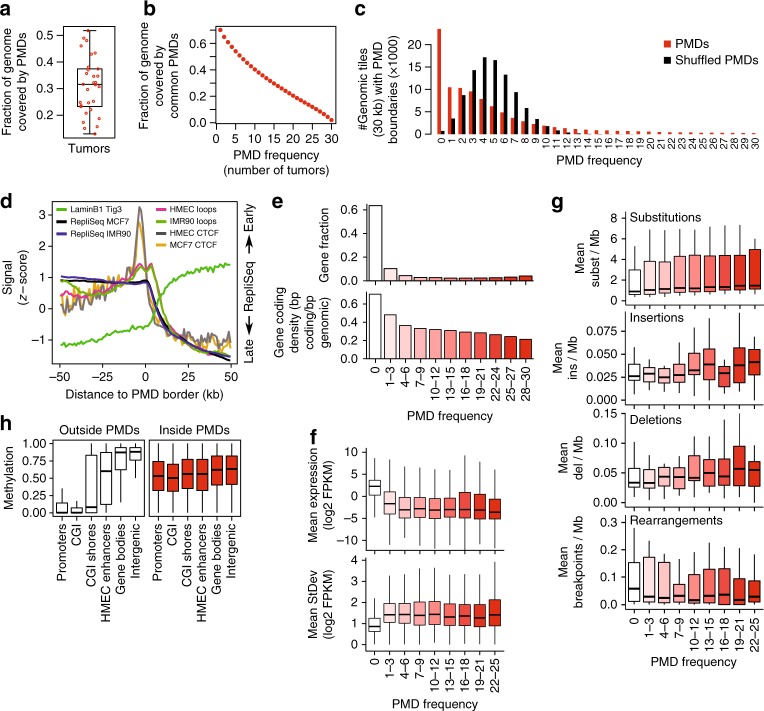
Fig. 2.
Characterization of breast cancer PMDs. a Fraction of the genome covered by PMDs. Each dot represents one tumor sample, the boxplot summarizes this distribution. b Fraction of the genome covered by PMDs that are common between breast tumors. PMD frequency: the number of tumors in which a genomic region or gene is a PMD. c Breast cancer PMDs are not distributed randomly over the genome. The genome was dissected into 30-kb tiles, PMD frequency (number of boundaries) was calculated for each tile. The same analysis was done after shuffling the PMDs of each tumor sample. d Average profiles of LaminB23, repliSeq (DNA replication timing, ENCODE), 3D chromatin interaction loops (HiC27, and CTCF (ENCODE) over PMD borders. If available, data from the breast cancer cell line (MCF7) and mammary epithelial cells (HMEC) was used, otherwise data from fibroblasts (IMR90, Tig3) was used. e Gene distribution inside PMDs (top, as a fraction of all annotated genes; bottom, as gene coding density). f Gene expression inside PMDs. Gene expression (top) and standard deviation (bottom) for the 25 overlapping cases of our WGBS and the transcriptome cohorts17 was plotted as a function of PMD frequency. g Somatic mutations inside PMDs. Substitutions, insertions, deletions, and rearrangements were calculated for the 25 overlapping cases of our WGBS and the breast tumor full genomes cohorts15, and plotted as a function of PMD frequency. h Distribution of DNA methylation over functional genomic elements, inside and outside PMDs. CpGs were classified according PMD status and genomic elements, and the distribution of DNA methylation within each element was plotted. All boxplots in this figure represent the median and 25th and 75th percentiles, whiskers 1.5 times the interquartile range, outliers are not shown