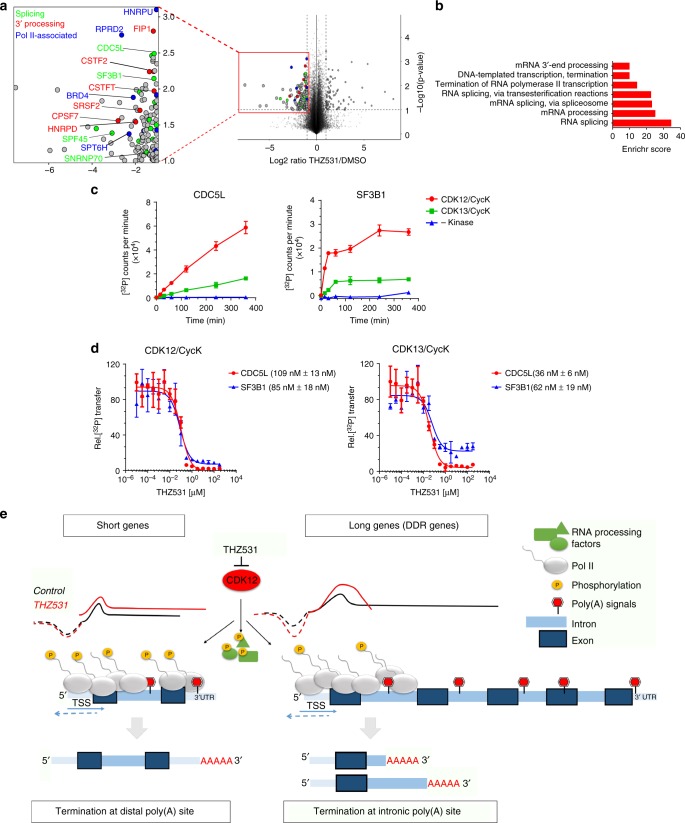
Fig. 6.
CDK12/13 phosphorylates RNA processing proteins. a Volcano plot of proteome-wide changes in phosphorylation site occupancy identified through SILAC analysis of NB cells treated with THZ531, 400 nM for 2 h. Expanded box shows selected co-transcriptional RNA processing proteins. b GO terms for candidate CDK12/13 substrates. c In vitro kinase assays of CDK12/CycK (red)-mediated and CDK13/CycK (green)-mediated phosphorylation of CDC5L (aa 370-505) and GST-SF3B1 (aa 113-462) at the indicated time points. A negative control measurement without kinase is shown in blue. Radioactive kinase reactions were performed with 0.2 µM CDK12/CycK or CDK13/CycK and 50 µM substrate protein, respectively. Data are reported as mean ± SD, n = 3. d Dose-response curves of THZ531 incubated with recombinant CDK12 (left) and CDK13 (right) protein and CDC5L (aa 370-505) and GST-SF3B1 (aa 113-462). Radioactive kinase reactions were performed after 30 min preincubation with increasing concentrations of THZ531. For all incubation time series, the counts per minute of the kinase activity measurements were normalized to the relative 32P transfer. Data are reported as mean ± SD; n = 3. IC50 values shown in parentheses. e Model of CDK12 as a regulator of pre-mRNA processing. CDK12 phosphorylates and thus likely stimulates the orchestrated action of RNA Pol II CTD and RNA processing proteins. CDK12 inhibition leads to a gene-length-dependent productive elongation defect associated with early termination through premature cleavage and polyadenylation (PCPA). Especially vulnerable to PCPA are long genes with a lower ratio of U1 snRNP binding to poly(A) sites, which include many of those involved in the DDR. Among short genes, including genes that normally terminate through stem-loop binding, CDK12 inhibition increases intron retention and leads to longer polyadenylated transcripts