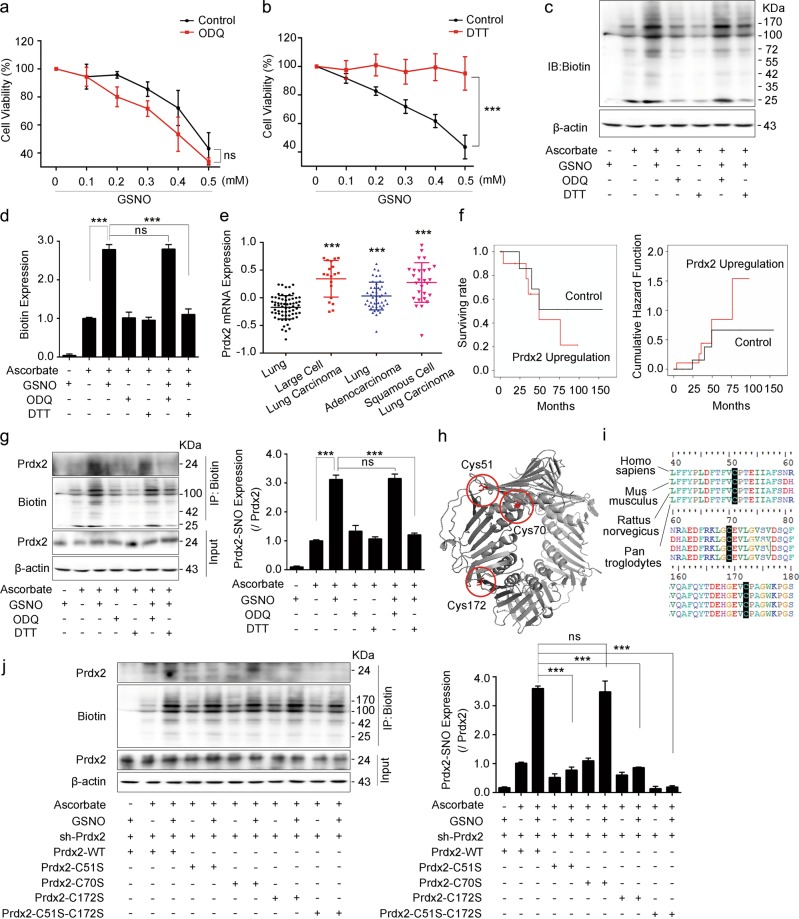
Fig. 2. GSNO promotes lung cancer cell apoptosis via Prdx2 nitrosylation.
a Effect of GSNO on cell viability with or without ODQ (25 μM) for 24 h (ns, not significant, using two-way ANOVA followed by Bonferroni’s multiple comparisons test). b Effect of GSNO on cell viability with or without DTT (0.5 mM) for 24 h (***P < 0.001, using two-way ANOVA followed by Bonferroni’s multiple comparisons test). c, d Detection of total protein nitrosylation by biotin-switch assay (***P < 0.001; ns, not significant, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). e The mRNA expression level of Prdx2 in different kinds of human lung cancer tissues and normal tissues (***P < 0.001, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). f Kaplan–Meier survival curves for cumulative survival rate and cumulative hazard function of cancer patients according to expression level of Prdx2 mRNA. g Detection of Prdx2 nitrosylation by biotin-switch assay (***P < 0.001; ns, not significant, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). h The 3D structure of Prdx2. The cysteine is shown in red. i Sequence alignment of Prdx2 in different species. j Expression of Prdx2 nitrosylation with GSNO treatment in different Prdx2 mutantions (***P < 0.001; ns, not significant, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). The data are expressed as the mean ± SD of three independent experiments