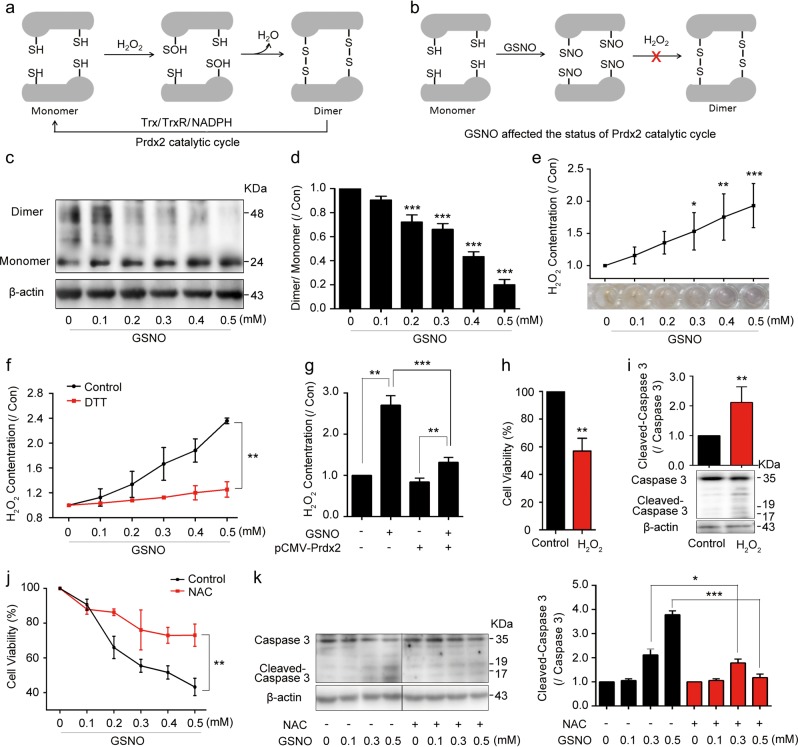
Fig. 3. GSNO nitrosylates Prdx2 to inhibit Prdx2 activity and causes H2O2 accumulation.
a Schematic representation of physiological Prdx2 catalytic mechanism of Prdx2. b GSNO affected the status of Prdx2 catalytic cycle. c, d Detection of Prdx2 dimmer/monomer status after treating A549 with GSNO for 24 h (***P < 0.001, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). e Detection of intracellular H2O2 after GSNO treatment for 24 h (*P < 0.05, **P < 0.01, ***P < 0.001, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). f Detection of intracellular H2O2 with or without DTT (**P < 0.01, using two-way ANOVA followed by Bonferroni’s multiple comparisons test). g Detected intracellular H2O2 in Prdx2-overexpression cells (**P < 0.01, ***P < 0.001, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). h Cell viability analysis of A549 cells with H2O2 (50 μM) treatment for 24 h (**P < 0.01, using unpaired two-tailed Student’s t-test). i Cleaved Caspase-3 expression in A549 cells with H2O2 treatment for 24 h (**P < 0.01, using unpaired two-tailed Student’s t-test). j Effect of GSNO on A549 cell viability with or without NAC (2 mM) (**P < 0.01, using two-way ANOVA followed by Bonferroni’s multiple comparisons test). k Cleaved-Caspase-3 expression in A549 cells with or without NAC (*P < 0.05, ***P < 0.001, using one-way ANOVA followed by Bonferroni’s multiple comparisons test). The data are expressed as the mean ± SD of three independent experiments