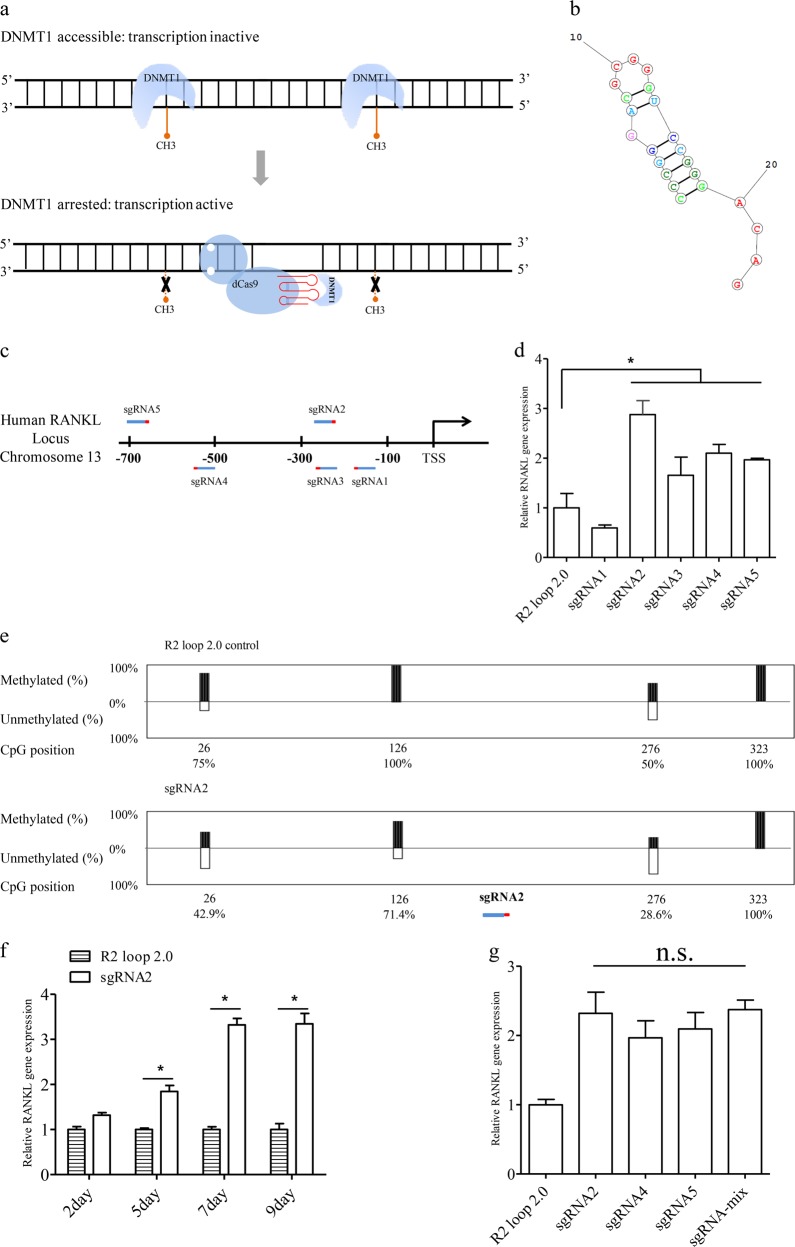
Fig. 1. CRISPR/dCas9-R2 based DNA demethylation system.
a Schematic description of demethylation of targeted DNA sequences by restricting the DNMT1 activity. The DNMT1 is recruited by the sgRNA-R2 stemloop, and the methyltransferase activity is therefore inhibited, resulting in the decrease of the DNA methylation level. b The RNA structure of the R2 stemloop. c Illustration of the five sgRNAs designed for targeting the promoter region of the human RANKL gene. The sgRNAs were shown in blue-red color, where the blue color represented the guide sequences and the red color represented the PAM region, and the sgRNAs located at about 700 bps upstream of the transcriptional start site of RANKL. d Determination of the abundances of RANKL mRNA in HEK-293T cells by quantitative RT-PCR assay. e Measurement of the percentages of methylated and unmethylated DNA in the RANKL promoter region by the bisulfate sequencing method. The presented data were first normalized to the reference group (i.e., the CRISPR/dCas9-R2 system using an sgRNA with no guide sequence), and the accurate value of methylation level was shown below each CpG site. f Assessment of the transcriptional level of RANKL gene by quantitative RT-PCR assay. Total RNA was prepared at the different post-transfection days from the HEK-293T cells (i.e., 2–9 days). g Determination of the additive effect of the CRISPR/dCas9-R2 system in upregulating gene expression by quantitative RT-PCR assay. Either individual sgRNA (i.e., RANKL sgRNA-2, 4 and 5) or a mixture of these sgRNAs resulted in a similar level of transcriptional activation