Maturity onset diabetes of the young (MODY) is a heterogenous, monogenic disease that is characterized by an autosomal dominant inheritance and an early disease onset, usually before 25 years of age (1). In contrast to late-onset forms of type 2 diabetes in which β-cell defects develop as a result of insulin resistance, MODY is caused by a primary defect in pancreatic β-cell function and impaired glucose-stimulated insulin secretion (2). Mutations in five genes are associated with different forms of MODY. The MODY2 gene encodes the glycolytic enzyme glucokinase that acts as the glucose sensor of the β cell and when inactivated, leads to impaired sensing of blood glucose levels (3). However, the majority of MODY forms are caused by mutations in transcription factors that are enriched in pancreatic β cells and include hepatocyte nuclear factors 1α (HNF-1α, MODY3), HNF-4α (MODY1), insulin promoter factor 1 (IPF-1/PDX-1, MODY4), and HNF-1β (MODY5) (see Table 1). These forms of diabetes share many pathophysiological features and are known to regulate the expression of genes such as the glucose transporter 2 (GLUT-2) and glycolytic genes that are essential for normal β-cell function (4–6). Studies performed in extrapancreatic tissues such as the liver, gut, and visceral endoderm have shown that HNFs form a hierarchical transcriptional network that regulates differentiation and metabolism in these cells (7–9). Two recent studies, including the paper by Boj et al. in this issue of PNAS (10), significantly advance our understanding of this regulatory circuit in pancreatic β cells.
Table 1.
Genetic classification of MODY subtypes
| Type | Gene | Frequency, %MODY families | Penetrance |
|---|---|---|---|
| MODY1 | HNF-4α | 2–4 | High |
| MODY2 | GCK | 11–63 | 100% |
| MODY3 | HNF-1α | 21–73 | High |
| MODY4 | IPF-1/PDX-1 | 1–4 | High |
| MODY5 | HNF-1β | 1–5 | High |
The transcriptional cascade of HNFs was first elucidated in hepatocytes where it was shown that HNF-4α, a member of the steroid hormone receptor superfamily, was an essential positive regulator of HNF-1α (11, 12). This was one of the first transcriptional hierarchies that was characterized in a mammalian system and shown to be essential for differentiation. Stable expression of HNF-4α in dedifferentiated hepatoma cells could induce re-expression of the endogenous HNF-1α gene. Studies using dedifferentiated hepatoma variants and intertypic cell hybrids revealed that the disappearance and re-expression of the hepatic phenotype are correlated with the abolition and expression, respectively, of HNF-4α and HNF-1α. Strong evidence showing that this functional hierarchy is also important in pancreatic β cells came from the finding that MODY could be caused by a mutation in the HNF-4α binding site of the HNF-1α promoter (13). The fact that this mutation occurred only in one allele but cosegregated with every diabetic individual in the affected pedigree suggests that HNF-4α is an essential activator of the HNF-1α gene. Boj et al. now present evidence that this relationship is not unidirectional but that HNF-1α is also an important activator of HNF-4α in the pancreas. Studies by Boj et al. (10) and Thomas et al. (14) independently describe a functional HNF-1 binding site in an alternative HNF-4α promoter, named P2, which is located ≈46 kb upstream of the previously identified P1 promoter of the HNF-4α gene (15). Both groups convincingly demonstrate that the P2 promoter is the major regulatory site for islet-specific HNF-4α transcription by the following genetic and biochemical evidence: (i) HNF-4α7 (the major islet isoform that is transcribed from the P2 promoter) was reduced >10-fold in pancreatic islets of HNF-1α-deficient mice compared with wild-type control animals (10), (ii) chromatin immunoprecipitation analysis indicated that HNF-1α occupies the HNF-4αP2 promoter in vivo (10), (iii) transient transfection in INS-1 cells showed that deletion of the HNF-1 binding site from the HNF-4αP2 led to a marked reduction in its transcriptional activity (14), and (iv) cotranfection of the P2 promoter with HNF-1α and HNF-1β led to a 2- and 4-fold increase, respectively, in transactivation of a reporter gene (14). Together, these data strongly suggest that HNF-4α and HNF-1α autoregulate each other and that a defect in this regulatory feed-forward loop is responsible for the common insulin secretory phenotype in subjects with MODY1 and MODY3.
Interestingly, the work by Thomas et al. (14) also reveals a link between HNF-4α and the MODY4 transcription factor PDX-1. They identified a mutation (-146T-C) in the HNF-4αP2 promoter of a large MODY family where a mutated PDX-1 binding site in the P2 promoter of the HNF-4α gene cosegregates with diabetes (logarithm of odds score of 3.25). This mutation led to decreased binding activity of PDX-1 to this site and reduced transcriptional activation. These data suggest then that PDX-1 is an important activator of HNF-4α7 expression in pancreatic β cells.
Understanding the regulation of the PDX-1 gene is of paramount importance because PDX-1 functions as a master regulator of pancreas development and β-cell differentiation (16). The PDX-1 gene is initially expressed in exocrine and endocrine pancreatic precursors but later becomes restricted mainly to β cells. Loss of PDX-1 expression leads to pancreatic agenesis and haploinsufficiency of the PDX-1 gene results in defects in glucose-stimulated insulin secretion in humans and mice (17–20). Elegant studies in which PDX-1 expression was specifically abolished in differentiated β cells of mice indicate that the PDX-1 gene in the adult islet is required for maintenance of the β-cell phenotype, including expression of insulin, GLUT-2, and glucokinase genes (20). Expression of PDX-1 in the adult islet is regulated by the feed-forward loop formed by the transcription factors HNF-1α and HNF-4α as well as by the forkead transcription factor HNF-3β (Foxa2) that may be positioned upstream in the transcriptional network (Fig. 1). Recent in vitro studies demonstrated that HNF-1α can bind to an evolutionarily conserved HNF-1 binding site in the PDX-1 promoter and activate its transcription (21). Furthermore, PDX-1 expression levels are reduced in islets of HNF-1α−/− mice, indicating that HNF-1α is required for normal PDX-1 expression in pancreatic β cells (22). Similarly HNF-3β, a member of the forkhead/winged helix family of transcription factors, has also been shown to bind to conserved regulatory elements in the mouse and human PDX-1 promoters and to activate their transcription (21, 23, 24).
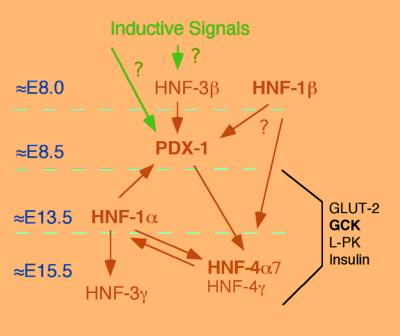
Figure 1.
Proposed model for the transcriptional network in pancreatic β cells. This diagram shows the transcriptional regulation of MODY genes that are essential for pancreatic islet function. Arrows show positive regulation. Bold symbols indicate MODY genes. The numbers on the left (E8.0–E15.5) indicate the approximate onset of gene expression during embryonic development.
The majority of our current understanding in the transcriptional control of islet-enriched genes involves positive regulators of gene expression. Only a few negative regulators of transcription have been studied in this context. Small heterodimer partner (SHP) is an orphan nuclear receptor that can heterodimerize with various nuclear hormone receptors, including HNF-4α, and inhibit their function (25, 26). A recent study showed that the expression of SHP is diminished in islets of HNF-1α−/− mice and this decrease is mediated secondarily by reduced expression of HNF-4α in these islets. SHP can regulate its own expression by inhibiting the function of its transcriptional regulator HNF-4α, thus forming a negative feedback loop (22). SHP therefore may serve as an important checkpoint to balance the activity of the islet transcriptional network. The precise physiological significance of this negative regulation, however, awaits further characterization in mutant SHP mouse models.
Preliminary data suggest that embryonic stem cells can differentiate into insulin-secreting cells.
Pancreatic islet-specific gene expression is to a large extent controlled at the transcriptional level by the binding of unique combinations of islet-enriched transcription factors to cis-acting sequences of target genes. The same transcription factors that are responsible for this combinatorial control also direct temporal gene expression profiles that are important for pancreatic development. Cell intrinsic and extrinsic factors initiate expression patterns that distinguish the prepancreatic region of the developing endoderm before the formation of the pancreatic buds (27). HNF-3β is a candidate for initiating endoderm patterning and may activate PDX-1. PDX-1 expression is first detected at ≈embryonic day 8.0 (E8.0) in the dorsal endoderm of the foregut/midgut junction before the appearance of early insulin and/or glucagon-expressing cells (16). These cells, however, lack the expression of other β-cell markers. Around E13, the number of endocrine cells start to increase and take on both morphological and molecular characteristics of a differentiated islet, including formation of cell clusters and increased insulin and glucagon expression per cell (28). The endocrine cells form well-organized islets by E18. Final differentiation of pancreatic islets occurs during the first week of life with the development of glucose-sensing capabilities and appropriate glucose-stimulated insulin secretion response (28). The study by Boj et al. (10) revealed that differentiation of islets in late development requires HNF-1α to maintain the expression of HNF-4α7, HNF-3γ, and GLUT-2. HNF-1α is expressed in many cell types of the developing pancreas at E13.5. At that time, GLUT-2, HNF-3γ, and HNF-4α7 are coexpressed with HNF-1α, and expression of these genes seem to be largely independent of Hnf-1α expression. However, the expression of GLUT-2, HNF-3γ, and HNF-4α7 is progressively reduced in HNF-1α−/− mice cells after E14 of embryonic development. This finding suggests that HNF-1α is not required to set up the early transcriptional program in β-cell differentiation but is necessary to maintain the expression pattern of the developing islet after ≈E14 and thus may define a novel stage in pancreatic β-cell development. This HNF-1α dependence is maintained throughout life in the mature islet (10, 22).
Why is a detailed molecular understanding of the transcriptional network of pancreatic β cell important? As we begin to understand the function and regulation of the many genes and signals involved in the differentiation process of β cells, it may be possible to use this knowledge to promote the generation of β cells in vitro for transplantation in type 1 diabetic patients. Preliminary data suggest that embryonic stem cells can be differentiated into insulin-secreting cells; however, these cells are not terminally differentiated as suggested by low insulin content and poor glucose-stimulated insulin secretion response (29). In addition, this knowledge may be exploited for in vivo regeneration of missing or malfunctioning β cells in type 2 diabetic subjects. Cells associated with the exocrine duct system of the pancreas may exhibit characteristics that are similar to multipotent progenitor cells that are found in the pancreatic epithelium of embryos (30, 31). A detailed knowledge of the transcriptional programs that guide β-cell differentiation in vivo will hopefully allow us to define defects in differentiation and manipulate those processes, either pharmacologically or by using gene delivery approaches. Furthermore, additional insights into the role of this transcriptional network in the early and late clinical stages of type 2 diabetes such as β-cell hypertrophy and β-cell decompensation may lead to novel approaches to delay or prevent β-cell failure and the development of diabetes.
Footnotes
See companion article on page 14481.
References
- 1.Hattersley A T. Diabetes Med. 1998;15:15–24. doi: 10.1002/(SICI)1096-9136(199801)15:1<15::AID-DIA562>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 2.Fajans S S, Bell G I, Polonsky K S. N Engl J Med. 2001;27:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 3.Matschinsky F M. Diabetes. 1990;39:647–652. doi: 10.2337/diab.39.6.647. [DOI] [PubMed] [Google Scholar]
- 4.Stoffel M, Duncan S A. Proc Natl Acad Sci USA. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parrizas M, Maestro M A, Boj S F, Paniagua A, Casamitjana R, Gomis R, Rivera F, Ferrer J. Mol Cell Biol. 2001;21:3234–3243. doi: 10.1128/MCB.21.9.3234-3243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoffel M. In: Frontiers in Diabetes, Vol 15: Molecular Pathogenesis of MODYs. Matschinsky F, Magnuson M A, editors. Basel: Karger; 2000. pp. 251–264. [Google Scholar]
- 7.Duncan S A, Navas M A, Dufort D, Rossant J, Stoffel M. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Ning G, Duncan S A. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 9.Shih D Q, Bussen M, Sehayek E, Ananthanarayanan M, Shneider B L, Suchy F J, Shefer S, Bollileni J S, Gonzalez F J, Breslow J L, Stoffel M. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 10.Boj S F, Párrizas M, Maestro M A, Ferrer J. Proc Natl Acad Sci USA. 2001;98:14481–14486. doi: 10.1073/pnas.241349398. . (First Published November 20, 2001; 10.1073/pnas.241349398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian J M, Schibler U. Genes Dev. 1991;5:2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- 12.Kuo C J, Conley P B, Chen L, Sladek F M, Darnell J E, Jr, Crabtree G R. Nature (London) 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 13.Gragnoli C, Lindner T, Cockburn B N, Kaisaki P J, Gragnoli F, Marozzi G, Bell G I. Diabetes. 1997;46:1648–1651. doi: 10.2337/diacare.46.10.1648. [DOI] [PubMed] [Google Scholar]
- 14.Thomas H, Jaschkowitz K, Frayling T M, Mitchell S M S, Roosen S, Lingott-Frieg A, Tack C J, Ellard S, Ryffel G U, Hattersley A T. Hum Mol Genet. 2001;10:2089–2097. doi: 10.1093/hmg/10.19.2089. [DOI] [PubMed] [Google Scholar]
- 15.Zhong W, Mirkovitch J, Darnell J E., Jr Mol Cell Biol. 1994;14:7276–7284. doi: 10.1128/mcb.14.11.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edlund H. Diabetes. 1998;47:1817–1823. doi: 10.2337/diabetes.47.12.1817. [DOI] [PubMed] [Google Scholar]
- 17.Stoffers A, Ferrer J, Clarke W L, Habener J F. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 18.Stoffers D A, Zinkin N T, Stanojevic V, Clarke W L, Habener J F. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson J, Carlson L, Edlund T, Edlund H. Nature (London) 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 20.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-Shushan E, Marshak S, Shoshkes M, Cerasi E, Melloul D. J Biol Chem. 2001;276:17533–17540. doi: 10.1074/jbc.M009088200. [DOI] [PubMed] [Google Scholar]
- 22.Shih D Q, Screenan S, Munoz K N, Philipson L, Pontoglio M, Yaniv M, Polonsky K S, Stoffel M. Diabetes. 2001;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- 23.Wu K L, Gannon M, Peshavaria M, Offield M F, Henderson E, Ray M, Marks A, Gamer L W, Wright C V, Stein R. Mol Cell Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerrish K, Gannon M, Shih D, Henderson E, Stoffel M, Wright C V, Stein R. J Biol Chem. 2000;275:3485–3494. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- 25.Seol W, Choi H S, Moore D D. Science. 1996;272:1336–1369. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y K, Dell H, Dowhan D H, Hadzopoulou-Cladaras M, Moore D D. Mol Cell Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grapin-Botton A, Melton D A. Trends Genet. 2000;16:124–145. doi: 10.1016/s0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- 28.Pictet R, Rutter W J. In: Handbook of Physiology. Steiner D F, Frenkel N, editors. Washington DC: Williams & Wilkins; 1972. , Section 7, Vol. 1, pp. 25–66. [Google Scholar]
- 29.Lumelsky N, Blondel O, Laeng P, Velasco I, Ravin R, McKay R. Science. 2001;292:1389–1394. doi: 10.1126/science.1058866. [DOI] [PubMed] [Google Scholar]
- 30.Bouwens L, Pipeleers D G. Diabetologia. 1998;41:629–633. doi: 10.1007/s001250050960. [DOI] [PubMed] [Google Scholar]
- 31.Trivedi N, Hollister-Lock J, Lopez-Avalos M D, O'Neil J J, Keegan M, Bonner-Weir S, Weir G C. Endocrinology. 2001;142:2115–2122. doi: 10.1210/endo.142.5.8162. [DOI] [PubMed] [Google Scholar]