Figure 1. The asPARP approach and a multiple sequence alignment of the human PARP family of proteins focusing on the D-loop.
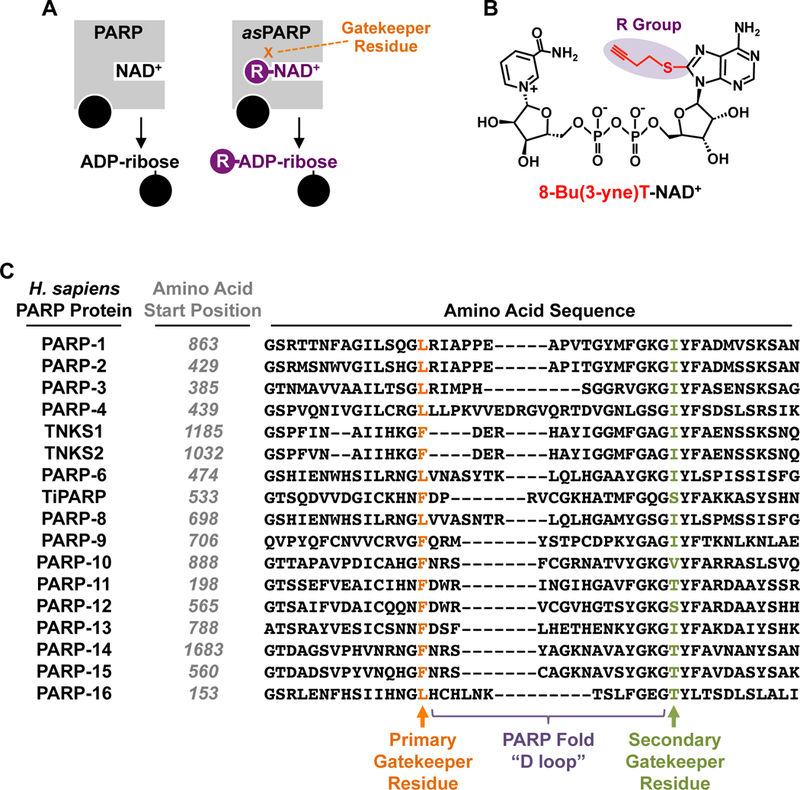
(A) Schematic representation of the asPARP approach using a single bi-functional and “clickable” NAD+ analog.
(B) Chemical structure of the clickable NAD+ analog 8-Bu(3-yne)T-NAD+.
(C) Multiple sequence alignment of a portion of the PARP catalytic domain, focusing on the D-loop, for all proteins in the human PARP family. The gatekeeper residues that confer NAD+ analog sensitivity are highlighted. For each PARP protein, the number of the first amino acid shown in the linear sequence of that PARP protein is indicated. “Primary Gatekeeper” refers to the amino acid in the PARP-1 catalytic domain where the most robust analog sensitivity was observed upon mutation to alanine using an automodification-based screen with a library of NAD+ analogs [12]. “Secondary Gatekeeper” refers to a second amino acid whose mutation also conferred analog sensitivity, but to a lesser extent and within a more constrained chemical space of NAD+ analogs [12]. These two sites are separated by the PARP “D loop,” but are located in proximity in three-dimensional space [12]. Although primary gatekeeper residue has been most useful for the asPARP approach to date, other NAD+ analogs screened in the future may show preferential activity with the secondary gatekeeper residue.
