Figure 4. Using asPARP methodologies for the discovery of PARP target proteins.
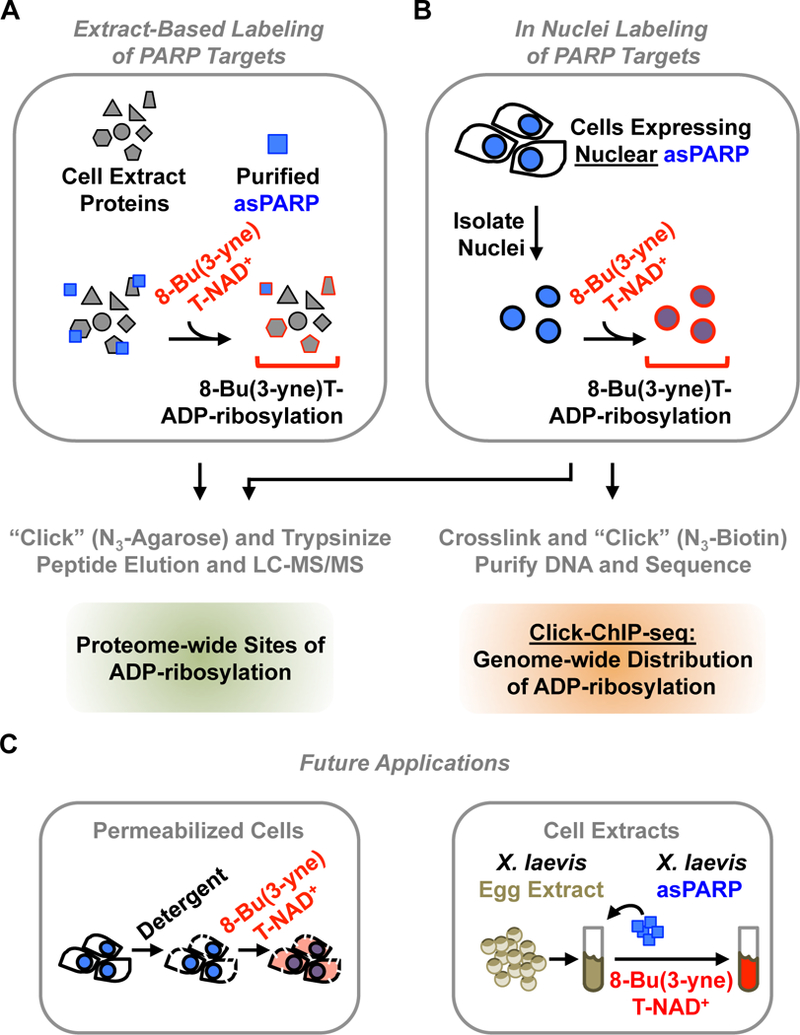
A schematic representation of different proven, as well as promising, applications of asPARP-dependent target labeling methodologies.
(A) In extract-based methods, a purified asPARP protein is used with a cellular extract and 8-Bu(3-yne)T-NAD+, as described in Figure 2. Sites of ADP-ribosylation on PARP target proteins can be identified by LC-MS/MS.
(B) For PARP proteins, which are expressed in the nucleus, in nuclei labeling is possible. Nuclei from nuclear asPARP-expressing cells are isolated and incubated with 8-Bu(3-yne)T-NAD+, stimulating spontaneous resumption of asPARP-driven 8-Bu(3-yne)T-ADP-ribosylation of nuclear PARP target proteins, as described in Figure 6. Sites of ADP-ribosylation on PARP target proteins can be identified by LC-MS/MS. Alternatively, sites of ADP-ribosylation across the genome can be determined by Click-ChIP-seq [12]. In click ChIP-seq, labeled nuclei are crosslinked with formaldehyde, clicked to azido-biotin (N3-biotin), and sonicated to shear the genomic DNA. This is followed by streptavidin-based purification of ADP-ribosylated chromatin and subsequent high throughput sequencing of the purified DNA.
(C) The asPARP technology may be developed further in the future to include more relevant or faithful biological contexts that recapitulate those that are found in vivo. (Left) Advances may include opportunities for delivering the clickable NAD+ analog directly into cells. This may entail careful permeabilization of cells using detergents, or perhaps even the development of new cell-permeable NAD+ analogs. (Right) Advances may also include opportunities to take advantage of complex biochemical extract preparations, such as the X. laevis egg extract system, where concerns such as dilution and unnatural buffer composition are less of an issue than with traditional extract preparation methods. Nearly all of the human PARP proteins have a X. laevis homolog with an identical primary gatekeeper residue. Furthermore, X. laevis egg extract preparations have been used with unparalleled success to reconstruct many dynamic biological processes [8], suggesting that the development of this system may offer an opportunity to uncover important PARP targets and molecular mechanisms that have remained conserved through evolution.
