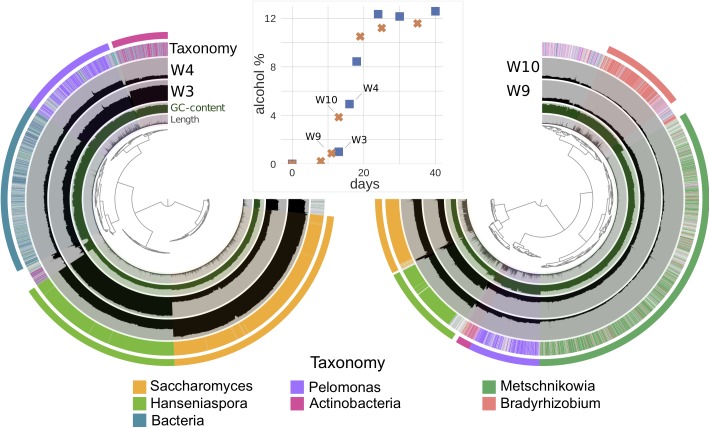
FIGURE 3.
The visualization of mean coverage of genome bins in samples W3 and W4 in vineyards 4 (left), and W9, W10 in vineyard 5 (right) using Anvi’o and their corresponding alcohol percentages during fermentation (middle; blue square: vineyard 4; orange cross: vineyard 5). GC-content, length of each contig, and taxonomy classified by Kaiju were displayed in adjunct layers. Each sample is represented in a separate layer, and each bar inside the sample layer corresponds to a datum computed for a given split, where contigs longer than 20k bp were divided to different splits. The outermost layer was the bin layer, where corresponding colors linked with the taxonomy of each bin. Vineyard 4 with 4333 contigs (minimum length: 2500 nucleotides, total nucleotides: 27.29 Mbp) that represented 4% of all contigs and 36% of all nucleotides found in the vineyard 4 contigs database (93,546 contigs and total nucleotides: 74.84 Mbp). Vineyard 5 with 5364 contigs (minimum length: 2500 nucleotides, total nucleotides: 53.01 Mbp) that represented 5% of all contigs and 50% of all nucleotides found in the vineyard 5 contigs database (92,948 contigs and total nucleotides: 105.44 Mb). The hierarchical clustering of contigs based on the sequence composition and their sample distribution were used for the dendrograms at the center of Anvi’o visualization. More bin details are shown in Supplementary Table S6.