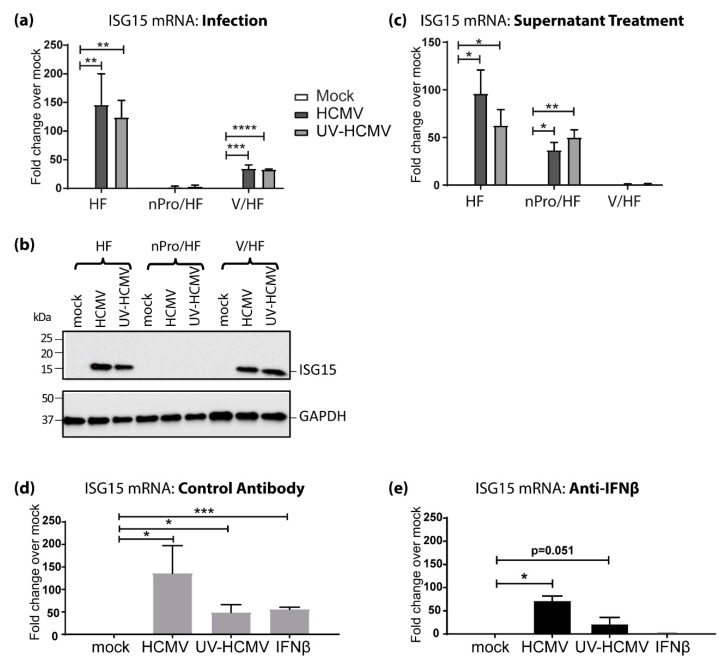
Figure 2.
IRF3-dependent, STAT1-independent regulation of ISG15 following HCMV infection (a) Primary HFs, nPro/HFs and V/HFs were infected at an MOI of 3 with HCMV or UV-HCMV and levels of ISG15 transcript were analyzed 6 h.p.i as in Figure 1. (b) Primary HFs, nPro/HFs and V/HFs were infected at an MOI of 3 with HCMV or UV-HCMV. Protein lysates were harvested at 24 h.p.i. and analyzed by immunoblot, staining for ISG15 and GAPDH. (c) Primary HFs, nPro/HFs and V/HFs were treated with filtered (0.1 μm pore size) supernatant from mock, HCMV or UV-HCMV infected primary HFs. RNA harvested at 6 h post-treatment was analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) as in Figure 1. RNA was extracted at 6 h.p.i. from primary HFs treated with 100 U of IFNβ or infected at a multiplicity of infection (MOI) of 3 with HCMV or UV-HCMV in the presence of (d) a control antibody or (e) IFNβ-neutralizing antibody and analyzed by qRT-PCR. Error bars indicate the standard error of the mean (SEM) and statistical significance was calculated using a Student’s two-tailed T-test. n = 3, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.