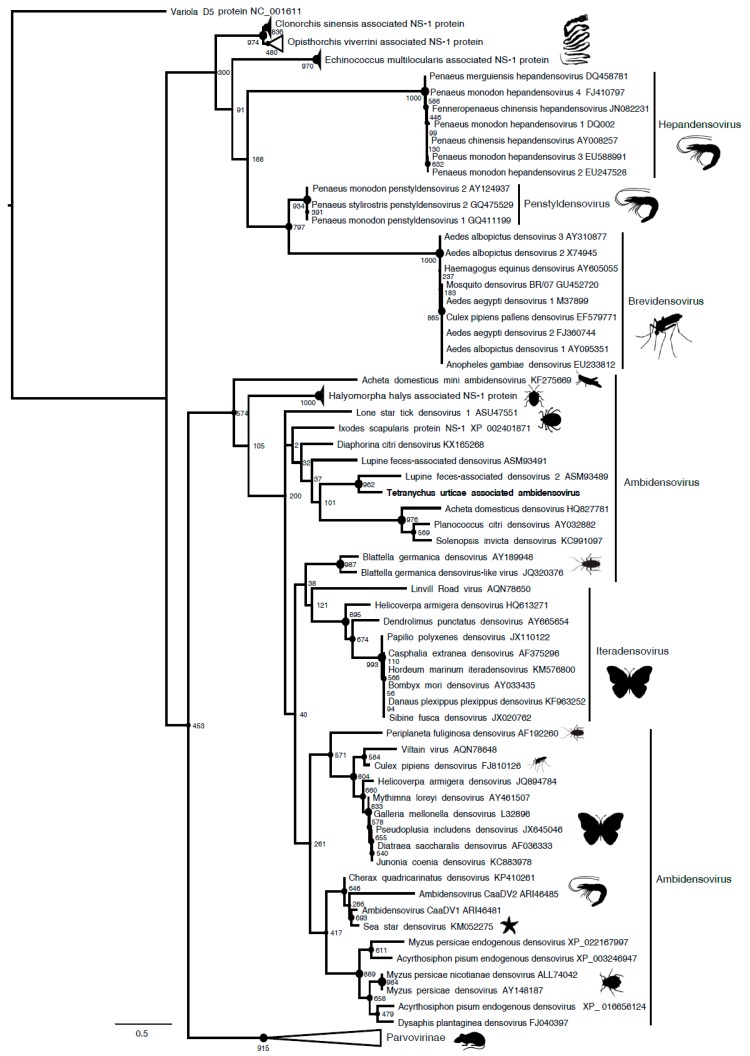
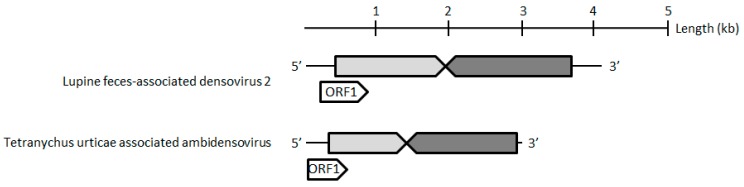
Figure 2.
Maximum Parvoviridae likelihood phylogenetic tree based on a part of the NS1 protein containing the SF3 domain, including 105 parvovirus sequences and Tetranychus urticae-associated ambidensovirus (in bold). The alignment of 155 amino acids in length was produced using MUSCLE 3.7 (16 iterations) and was ungapped by hand. The tree was rooted with the SF3 domain of the Variola virus D5 protein. Bootstrap values are indicated at each node. Scale bar corresponds to amino acid substitutions per site. Genera of the Parvoviridae family are indicated in brackets. Genomic organization of Tetranychus urticae-associated ambidensovirus is also indicated. Grey arrows and rectangles: predicted open reading frames (ORF), Light grey: putative NS; dark grey: putative capsid protein (CP or VP). Arrow: complete ORF.