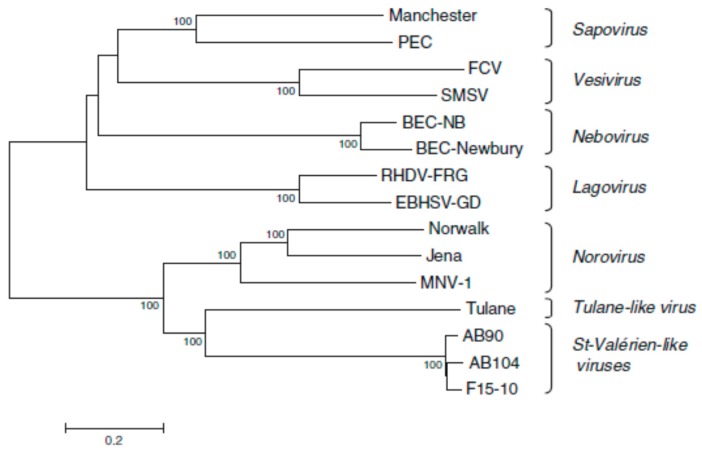
Figure 3.
Phylogenetic tree based on nucleotide sequences of complete calicivirus genomes differentiating Valovirus as a separate genus. The tree was constructed by the neighbour-joining method MEGA 3.1. The confidence values at the branch points are based on 1000 bootstrap analyses. The calibration bar indicates distance expressed as nt substitutions per site. Abbreviations: BEC-NB, bovine enteric calicivirus NB/80/US; BEC-Newbury, bovine enteric calicivirus Newbury; EBHSV-GD, European brown hare syndrome virus GD strain; FCV, feline calicivirus; Jena, bovine enteric norovirus strain Jena; Manchester, human sapovirus Manchester; MNV-1, mouse norovirus 1; Norwalk, Norwalk virus; PEC, porcine enteric calicivirus; RHDV-FRG, rabbit haemorrhagic disease virus Germany; SMSV, San Miguel Sea Lion Virus; Tulane, Tulane virus; and St Valérien, St Valérien strains AB90, AB104, F15-10. From reference [25], with permission of authors and publisher.