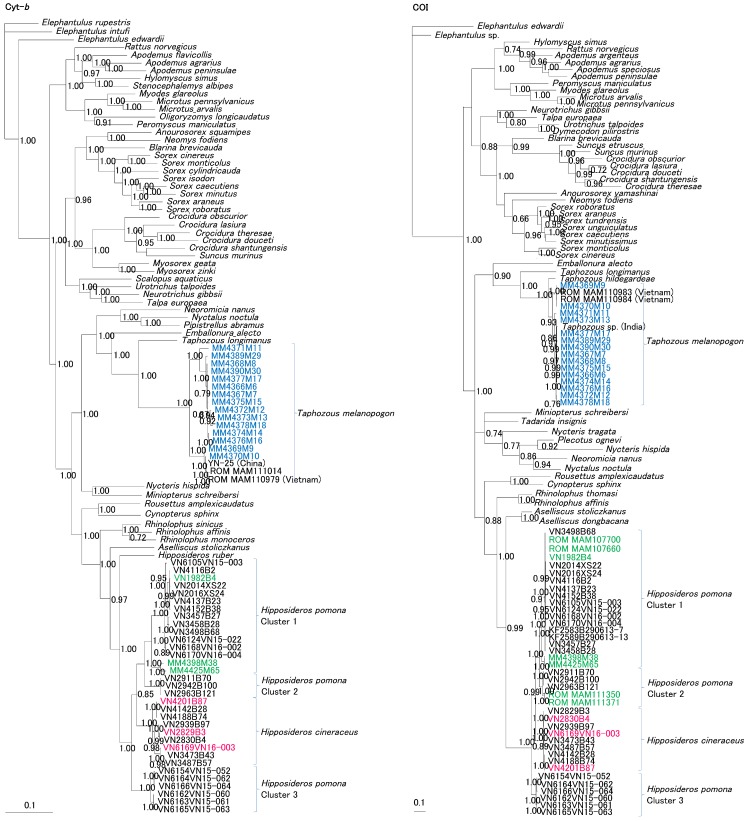
Figure 4.
Bayesian phylogenetic trees of the host organisms of mobatviruses reconstructed from the alignments of 1140-nucleotide cytochrome b (Cyt b) and 1545-nucleotide cytochrome oxidase I (COI) gene sequences. Taphozous melanopogon collected in Myanmar (blue), XSV-positive Hipposideros pomona (green) in Myanmar and XSV-positive Hipposideros cineraceus (red) in Vietnam are shown. Numbers at the nodes indicate posterior probability values (>0.7) based on 150,000 trees: two replicate Markov chain Monte Carlo runs, consisting of six chains of 10 million generations, each sampled every 100 generations, with a burn-in of 25,000 (25%). Scale bars indicate nucleotide substitutions per site. Gene accession numbers are listed in Table S4.