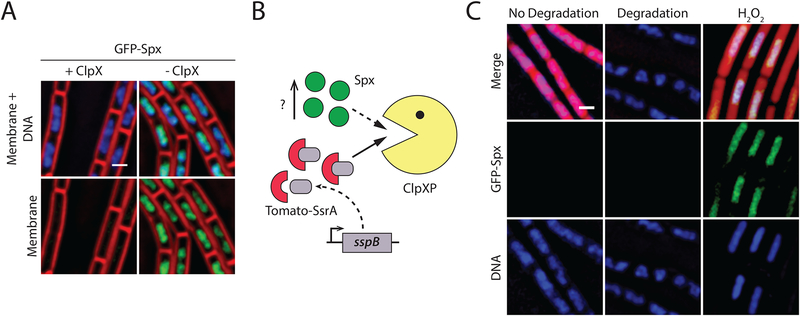
Figure 2. Degradation of an abundant protein does not affect turnover of a native ClpXP substrate.
A. Fluorescence microscopy of a strain carrying a GFP-Spx fusion that expresses ClpXP under the control of an IPTG-inducible promoter (BER0552), grown in the presence (left, +ClpXP) or absence (right, -ClpXP) of 1mM IPTG. When ClpX is limiting, GFP-Spx accumulates. Membranes are stained with FM4–64 (red) and DNA with DAPI (blue). Scale bar, 1μm.
B. Inducing degradation of abundant ssrA*-tagged proteins should cause accumulation of GFP-Spx if ClpXP is saturated.
C. Fluorescence microscopy of a strain carrying a GFP-Spx fusion, producing the red fluorescent protein, tomato, from a strong promoter and SspBEc from a xylose-inducible promoter (BER0577), grown in the absence (left, no degradation) or in the presence (middle, degradation) of 1% xylose, or with 2mM H2O2 (right) as a positive control. Imaging was performed 2 hours after treatment. Inducing degradation does not yield an accumulation of GFP-Spx, but treatment with hydrogen peroxide, which is known to evoke the Spx response, does. Membranes are stained with FM4–64 (red) and DNA with DAPI (blue). Scale bar, 1 μm.