Figure 6. Dynamic characteristics of GMPR complexes revealed by high resolution 31P field cycling NMR relaxometry.
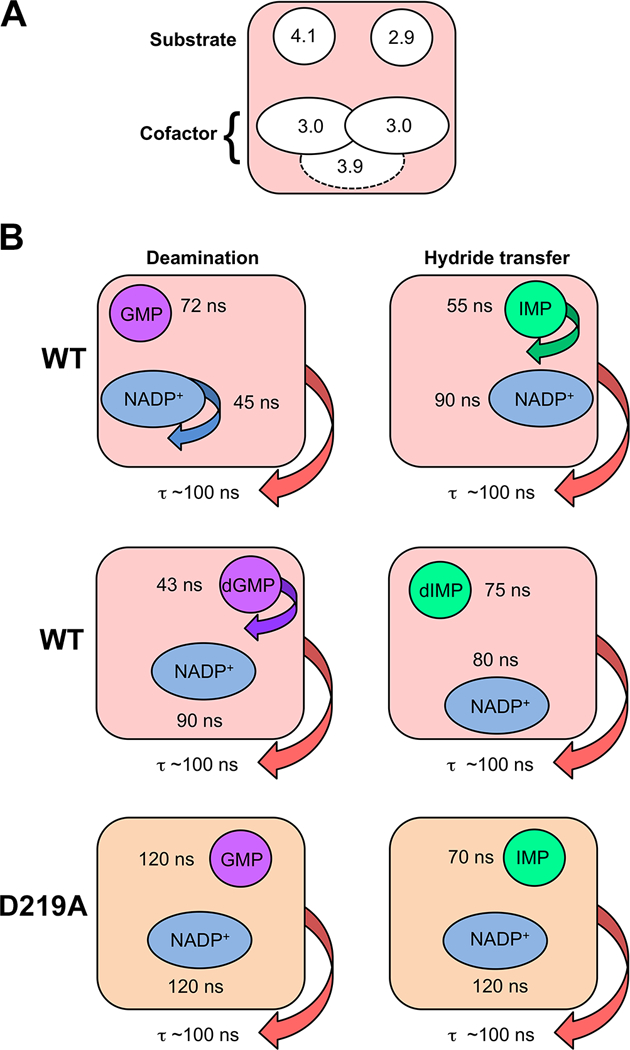
(A) Multiple binding modes for substrate and cofactors. Two distinct binding modes are observed for IMP and GMP in their catalytically active NADP+ complexes. The cofactor also has two distinct binding modes in these complexes as shown by the effects of D2O on relaxation, albeit with similar 31P-proton relaxer distances. These binding modes are indicated by the positions of the two ellipses. When the binding mode cannot be distinguished by the values of τ/R0, the ellipse is positioned in the center. A third conformation, indicated by the dashed ellipse, is observed in the dIMP-cofactor complex. The extrapolated 31P-proton relaxer distances are show in Å. (B) Distinct dynamic states are observed in wild-type enzyme and D219A and with ribose and deoxyribose substrates as evidenced by values of τ.
