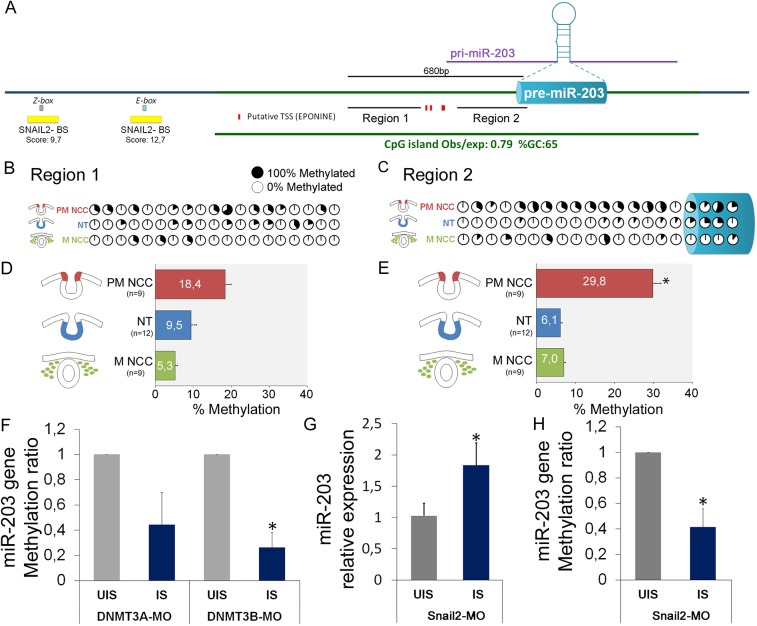
Fig. 5.
SNAIL2 and DNMT3B are required for DNA methylation of the miR-203 locus on premigratory NCC. (A) miR-203 genomic context. The CpG island content (green bar) for the miR-203 gene was identified using the UCSC genome browser (genome.ucsc.edu/) and Ensembl (www.ensembl.org/). The putative transcriptional start sites (TSS) (Tobiasson et al., 2017) were obtained using Eponine (www.sanger.ac.uk/science/tools/eponine). The putative binding sites for SNAIL2 were mapped using JASPAR 2018 (jaspar.genereg.net/). We also show the two regions used in our bisulfite sequencing and the pre-miR-203 accordingly to miRbase (www.mirbase.org/). (B,C) Bisulfite sequencing profiles of CpG site methylation in region 1 (B) and region 2 (C) were analyzed on pre-migratory NCCs (PM-NCCs), migratory NCCs (M-NCCs) and ventral neural tube (NT). The percentages of methylated CpG sites are shown with filled (100% methylated) and open (0% methylated) circles. (D,E) Bar graphs show the total percentages of methylated CpG sites on the different regions for the three analyzed samples in region 1 (D) and in region 2 (E). We found a higher percentage of methylated CpGs in PM-NCCs compared with other samples. Number in brackets indicate the number of analyzed sequences. *P<0.05 (ANOVA). (F) Morpholino-mediated loss of DNMT3A (DNMT3A-MO) and DNMT3B (DNMT3B-MO) function results in a reduction of methylated CpG sites on the injected side (IS) compared with the uninjected side (UIS) of the same group of embryos. Independent clones (n=10) were sequence out of two samples containing six half-electroporated embryos. (G,H) Morpholino-mediated loss of SNAIL2 (SNAIL2-MO) function maintains an elevated level of miR-203 expression (G) and reduced CpGs methylation in region 2 (H) on the IS compared with the UIS of the same group of embryos (n=6). *P<0.05 (Student's t-test).